Photoactivation Mechanism, Timing of Protein Secondary Structure Dynamics and Carotenoid Translocation in the Orange Carotenoid Protein
- PMID: 30511841
- PMCID: PMC6331140
- DOI: 10.1021/jacs.8b11373
Photoactivation Mechanism, Timing of Protein Secondary Structure Dynamics and Carotenoid Translocation in the Orange Carotenoid Protein
Abstract
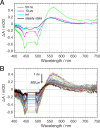
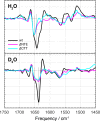
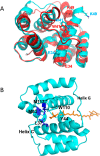
The orange carotenoid protein (OCP) is a two-domain photoactive protein that noncovalently binds an echinenone (ECN) carotenoid and mediates photoprotection in cyanobacteria. In the dark, OCP assumes an orange, inactive state known as OCPO; blue light illumination results in the red active state, known as OCPR. The OCPR state is characterized by large-scale structural changes that involve dissociation and separation of C-terminal and N-terminal domains accompanied by carotenoid translocation into the N-terminal domain. The mechanistic and dynamic-structural relations between photon absorption and formation of the OCPR state have remained largely unknown. Here, we employ a combination of time-resolved UV-visible and (polarized) mid-infrared spectroscopy to assess the electronic and structural dynamics of the carotenoid and the protein secondary structure, from femtoseconds to 0.5 ms. We identify a hereto unidentified carotenoid excited state in OCP, the so-called S* state, which we propose to play a key role in breaking conserved hydrogen-bond interactions between carotenoid and aromatic amino acids in the binding pocket. We arrive at a comprehensive reaction model where the hydrogen-bond rupture with conserved aromatic side chains at the carotenoid β1-ring in picoseconds occurs at a low yield of <1%, whereby the β1-ring retains a trans configuration with respect to the conjugated π-electron chain. This event initiates structural changes at the N-terminal domain in 1 μs, which allow the carotenoid to translocate into the N-terminal domain in 10 μs. We identified infrared signatures of helical elements that dock on the C-terminal domain β-sheet in the dark and unfold in the light to allow domain separation. These helical elements do not move within the experimental range of 0.5 ms, indicating that domain separation occurs on longer time scales, lagging carotenoid translocation by at least 2 decades of time.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Demmigadams B. Biochim. Biophys. Acta, Bioenerg. 1990, 1020, 1.10.1016/0005-2728(90)90088-L. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

