The Modes of Action of MARTX Toxin Effector Domains
- PMID: 30513802
- PMCID: PMC6315884
- DOI: 10.3390/toxins10120507
The Modes of Action of MARTX Toxin Effector Domains
Abstract
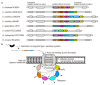
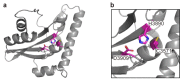
Many Gram-negative bacterial pathogens directly deliver numerous effector proteins from the bacterium to the host cell, thereby altering the target cell physiology. The already well-characterized effector delivery systems are type III, type IV, and type VI secretion systems. Multifunctional autoprocessing repeats-in-toxin (MARTX) toxins are another effector delivery platform employed by some genera of Gram-negative bacteria. These single polypeptide exotoxins possess up to five effector domains in a modular fashion in their central regions. Upon binding to the host cell plasma membrane, MARTX toxins form a pore using amino- and carboxyl-terminal repeat-containing arms and translocate the effector domains into the cells. Consequently, MARTX toxins affect the integrity of the host cells and often induce cell death. Thus, they have been characterized as crucial virulence factors of certain human pathogens. This review covers how each of the MARTX toxin effector domains exhibits cytopathic and/or cytotoxic activities in cells, with their structural features revealed recently. In addition, future directions for the comprehensive understanding of MARTX toxin-mediated pathogenesis are discussed.
Keywords: MARTX toxin; bacterial protein toxin; effector domain; host–microbe interaction; host–pathogen interaction.
Conflict of interest statement
The author declares no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures






Similar articles
-
Distinct roles of the repeat-containing regions and effector domains of the Vibrio vulnificus multifunctional-autoprocessing repeats-in-toxin (MARTX) toxin.mBio. 2015 Mar 31;6(2):e00324-15. doi: 10.1128/mBio.00324-15. mBio. 2015. PMID: 25827415 Free PMC article.
-
MARTX toxins as effector delivery platforms.Pathog Dis. 2015 Dec;73(9):ftv092. doi: 10.1093/femspd/ftv092. Epub 2015 Oct 15. Pathog Dis. 2015. PMID: 26472741 Free PMC article. Review.
-
Multifunctional-autoprocessing repeats-in-toxin (MARTX) Toxins of Vibrios.Microbiol Spectr. 2015 Jun;3(3):10.1128/microbiolspec.VE-0002-2014. doi: 10.1128/microbiolspec.VE-0002-2014. Microbiol Spectr. 2015. PMID: 26185092 Free PMC article. Review.
-
Coordinated delivery and function of bacterial MARTX toxin effectors.Mol Microbiol. 2018 Jan;107(2):133-141. doi: 10.1111/mmi.13875. Epub 2017 Dec 5. Mol Microbiol. 2018. PMID: 29114985 Free PMC article. Review.
-
Cytotoxicity of the Vibrio vulnificus MARTX toxin effector DUF5 is linked to the C2A subdomain.Proteins. 2014 Oct;82(10):2643-56. doi: 10.1002/prot.24628. Epub 2014 Jun 26. Proteins. 2014. PMID: 24935440 Free PMC article.
Cited by
-
A deep-sea bacterium related to coastal marine pathogens.Environ Microbiol. 2021 Sep;23(9):5349-5363. doi: 10.1111/1462-2920.15629. Epub 2021 Jun 15. Environ Microbiol. 2021. PMID: 34097814 Free PMC article.
-
The Vibrio cholerae MARTX toxin silences the inflammatory response to cytoskeletal damage before inducing actin cytoskeleton collapse.Sci Signal. 2020 Jan 14;13(614):eaaw9447. doi: 10.1126/scisignal.aaw9447. Sci Signal. 2020. PMID: 31937566 Free PMC article.
-
Biantennary N-glycans As Receptors for MARTX Toxins in Vibrio Pathogenesis.bioRxiv [Preprint]. 2024 Sep 12:2024.09.12.611726. doi: 10.1101/2024.09.12.611726. bioRxiv. 2024. PMID: 39314294 Free PMC article. Preprint.
-
Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.Proc Natl Acad Sci U S A. 2019 Sep 3;116(36):18031-18040. doi: 10.1073/pnas.1905095116. Epub 2019 Aug 19. Proc Natl Acad Sci U S A. 2019. PMID: 31427506 Free PMC article.
-
Vibrio vulnificus RtxA1 cytotoxin targets filamin A to regulate PAK1- and MAPK-dependent cytoskeleton reorganization and cell death.Emerg Microbes Infect. 2019;8(1):934-945. doi: 10.1080/22221751.2019.1632153. Emerg Microbes Infect. 2019. PMID: 31237474 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

