Genetic Hierarchy of Acute Myeloid Leukemia: From Clonal Hematopoiesis to Molecular Residual Disease
- PMID: 30513905
- PMCID: PMC6321602
- DOI: 10.3390/ijms19123850
Genetic Hierarchy of Acute Myeloid Leukemia: From Clonal Hematopoiesis to Molecular Residual Disease
Abstract
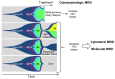
Recent advances in the field of cancer genome analysis revolutionized the picture we have of acute myeloid leukemia (AML). Pan-genomic studies, using either single nucleotide polymorphism arrays or whole genome/exome next generation sequencing, uncovered alterations in dozens of new genes or pathways, intimately connected with the development of leukemia. From a simple two-hit model in the late nineties, we are now building clonal stories that involve multiple unexpected cellular functions, leading to full-blown AML. In this review, we will address several seminal concepts that result from these new findings. We will describe the genetic landscape of AML, the association and order of events that define multiple sub-entities, both in terms of pathogenesis and in terms of clinical practice. Finally, we will discuss the use of this knowledge in the settings of new strategies for the evaluation of measurable residual diseases (MRD), using clone-specific multiple molecular targets.
Keywords: acute myeloid leukemia; clonal hematopoiesis; genetic hierarchy; molecular residual disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. - DOI - PubMed
-
- Cancer Genome Atlas Research Network. Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A.G., Hoadley K., Triche T.J., Laird P.W., et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

