Matrix association region/scaffold attachment region (MAR/SAR) sequence: its vital role in mediating chromosome breakages in nasopharyngeal epithelial cells via oxidative stress-induced apoptosis
- PMID: 30514321
- PMCID: PMC6278157
- DOI: 10.1186/s12867-018-0116-5
Matrix association region/scaffold attachment region (MAR/SAR) sequence: its vital role in mediating chromosome breakages in nasopharyngeal epithelial cells via oxidative stress-induced apoptosis
Abstract
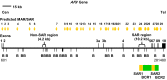
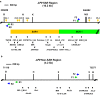
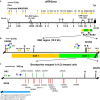
Background: Oxidative stress is known to be involved in most of the aetiological factors of nasopharyngeal carcinoma (NPC). Cells that are under oxidative stress may undergo apoptosis. We have previously demonstrated that oxidative stress-induced apoptosis could be a potential mechanism mediating chromosome breakages in nasopharyngeal epithelial cells. Additionally, caspase-activated DNase (CAD) may be the vital player in mediating the chromosomal breakages during oxidative stress-induced apoptosis. Chromosomal breakage occurs during apoptosis and chromosome rearrangement. Chromosomal breakages tend to cluster in certain regions, such as matrix association region/scaffold attachment region (MAR/SAR). We hypothesised that oxidative stress-induced apoptosis may result in chromosome breaks preferentially at the MAR/SAR sites. The AF9 gene at 9p22 was targeted in this study because 9p22 is a deletion site commonly found in NPC.
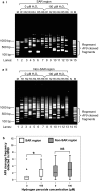
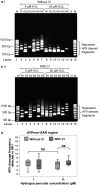
Results: By using MAR/SAR recognition signature (MRS), potential MAR/SAR sites were predicted in the AF9 gene. The predicted MAR/SAR sites precisely match to the experimentally determined MAR/SARs. Hydrogen peroxide (H2O2) was used to induce apoptosis in normal nasopharyngeal epithelial cells (NP69) and NPC cells (HK1). Nested inverse polymerase chain reaction was employed to identify the AF9 gene cleavages. In the SAR region, the gene cleavage frequency of H2O2-treated cells was significantly higher than that of the non-treated cells. A few chromosomal breakages were detected within the AF9 region which was previously found to be involved in the mixed lineage leukaemia (MLL)-AF9 translocation in an acute lymphoblastic leukaemia patient. As for the non-SAR region, no significant difference in the gene cleavage frequency was found between the untreated control and H2O2-treated cells. Furthermore, H2O2-induced cleavages within the SAR region were reduced by caspase-3 inhibitor, which indirectly inhibits CAD.
Conclusions: These results reaffirm our previous findings that oxidative stress-induced apoptosis could be one of the potential mechanisms underlying chromosome breakages in nasopharyngeal epithelial cells. MAR/SAR may play a vital role in defining the location of chromosomal breakages mediated by oxidative stress-induced apoptosis, where CAD is the major nuclease.
Keywords: AF9 gene; Apoptosis; H2O2; MAR/SAR; NPC; Oxidative stress.
Figures


References
-
- Raab-Traub N. Epstein–Barr virus and nasopharyngeal carcinoma. Semin Cancer Biol. 1992;3:297–307. - PubMed
-
- Armstrong RW, Armstrong MJ, Yu MC, Henderson BE. Salted fish and inhalants as risk factors for nasopharyngeal carcinoma in Malaysian Chinese. Cancer Res. 1983;43:2967–2970. - PubMed
-
- Huang DP, Ho JH, Saw D, Teoh TB. Carcinoma of the nasal and paranasal regions in rats fed Cantonese salted marine fish. IARC Sci Publ. 1978;20:315–328. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

