Mouse ICM Organoids Reveal Three-Dimensional Cell Fate Clustering
- PMID: 30514631
- PMCID: PMC6341222
- DOI: 10.1016/j.bpj.2018.11.011
Mouse ICM Organoids Reveal Three-Dimensional Cell Fate Clustering
Abstract
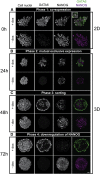
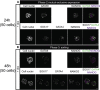
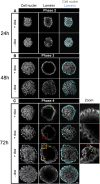
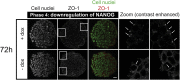
During mammalian preimplantation, cells of the inner cell mass (ICM) adopt either an embryonic or an extraembryonic fate. This process is tightly regulated in space and time and has been studied previously in mouse embryos and embryonic stem cell models. Current research suggests that cell fates are arranged in a salt-and-pepper pattern of random cell positioning or a spatially alternating pattern. However, the details of the three-dimensional patterns of cell fate specification have not been investigated in the embryo nor in in vitro systems. We developed ICM organoids as a, to our knowledge, novel three-dimensional in vitro stem cell system to model mechanisms of fate decisions that occur in the ICM. ICM organoids show similarities to the in vivo system that arise regardless of the differences in geometry and total cell number. Inspecting ICM organoids and mouse embryos, we describe a so far unknown local clustering of cells with identical fates in both systems. These findings are based on the three-dimensional quantitative analysis of spatiotemporal patterns of NANOG and GATA6 expression in combination with computational rule-based modeling. The pattern identified by our analysis is distinct from the current view of a salt-and-pepper pattern. Our investigation of the spatial distributions both in vivo and in vitro dissects the contributions of the different parts of the embryo to cell fate specifications. In perspective, our combination of quantitative in vivo and in vitro analyses can be extended to other mammalian organisms and thus creates a powerful approach to study embryogenesis.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Chazaud C., Yamanaka Y., Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell. 2006;10:615–624. - PubMed
-
- Guo G., Huss M., Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 2010;18:675–685. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

