Microbiota and Pathogen Proteases Modulate Type III Secretion Activity in Enterohemorrhagic Escherichia coli
- PMID: 30514785
- PMCID: PMC6282197
- DOI: 10.1128/mBio.02204-18
Microbiota and Pathogen Proteases Modulate Type III Secretion Activity in Enterohemorrhagic Escherichia coli
Abstract
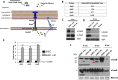
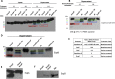
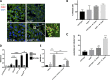
Enteric pathogens have complex interactions with the gut microbiota. Most of what is known about them has focused on microbiota-derived metabolites or small molecules that serve as nutrients and/or signals to aid in growth or transcriptionally regulate virulence gene expression. A common virulence strategy is to express a type III secretion system (T3SS), which is a molecular syringe deployed by many Gram-negative pathogens to hijack host cell function. Enterohemorrhagic Escherichiacoli (EHEC) requires its T3SS to colonize the intestinal tract and cause disease. Here we report that a prominent member of the intestinal microbiota, Bacteroides thetaiotamicron (Bt), secretes proteases that cleave the translocon of the T3SS of EHEC to enhance effector translocation into host cells. This is in contrast from an endogenous protease from EHEC itself (namely, EspP) that cleaves the translocon protein EspB in a different site to limit effector translocation. The EspB protein forms the T3SS pore in mammalian cells, and pore proteins are conserved in the T3SSs from several pathogens. This is the first demonstration of a commensal species directly processing a pathogen's T3SS, posing a new paradigm for how the microbiota can influence the severity of disease caused by bacterial pathogens. Because T3SSs are employed by many pathogens, this phenomenon has broad implications to commensal-pathogen relationships.IMPORTANCE The gut microbiota is usually regarded as providing colonization resistance against enteric pathogens. However, some pathogens evolved to thrive with the aid of certain members of the microbiota. Several Gram-negative bacteria employ type three secretion systems (T3SSs), which are molecular syringes that deliver effector proteins to host cells, hijacking host cell function. Here we show that the T3SS of enterohemorrhagic E. coli (EHEC) is cleaved by self and microbiota-derived proteases. Self-cleavage limits effector translocation, while cleavage by the microbiota member Bacteroides thetaiotamicron (Bt) exacerbates effector translocation and lesion formation on epithelial cells.
Keywords: Bacteroides; EspP; enterohemorrhagic E. coli (EHEC); microbiota; type three secretion.
Copyright © 2018 Cameron et al.
Figures

Comment in
-
Cuts Both Ways: Proteases Modulate Virulence of Enterohemorrhagic Escherichia coli.mBio. 2019 Feb 26;10(1):e00115-19. doi: 10.1128/mBio.00115-19. mBio. 2019. PMID: 30808700 Free PMC article.
References
-
- Tuttle J, Gomez T, Doyle MP, Wells JG, Zhao T, Tauxe RV, Griffin PM. 1999. Lessons from a large outbreak of Escherichia coli O157:H7 infections: insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol Infect 122:185–192. doi:10.1017/S0950268898001976. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

