Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex
- PMID: 30514815
- PMCID: PMC6305006
- DOI: 10.1073/pnas.1806438115
Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex
Abstract
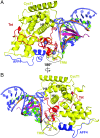
Promoter-proximal pausing by RNA polymerase II (Pol II) is a key regulatory step in human immunodeficiency virus-1 (HIV-1) transcription and thus in the reversal of HIV latency. By binding to the nascent transactivating response region (TAR) RNA, HIV-1 Tat recruits the human super elongation complex (SEC) to the promoter and releases paused Pol II. Structural studies of TAR interactions have been largely focused on interactions between the TAR bulge and the arginine-rich motif (ARM) of Tat. Here, the crystal structure of the TAR loop in complex with Tat and the SEC core was determined at a 3.5-Å resolution. The bound TAR loop is stabilized by cross-loop hydrogen bonds. It makes structure-specific contacts with the side chains of the Cyclin T1 Tat-TAR recognition motif (TRM) and the zinc-coordinating loop of Tat. The TAR loop phosphate backbone forms electrostatic and VDW interactions with positively charged side chains of the CycT1 TRM. Mutational analysis showed that these interactions contribute importantly to binding affinity. The Tat ARM was present in the crystallized construct; however, it was not visualized in the electron density, and the TAR bulge was not formed in the RNA construct used in crystallization. Binding assays showed that TAR bulge-Tat ARM interactions contribute less to TAR binding affinity than TAR loop interactions with the CycT1 TRM and Tat core. Thus, the TAR loop evolved to make high-affinity interactions with the TRM while Tat has three roles: scaffolding and stabilizing the TRM, making specific interactions through its zinc-coordinating loop, and making electrostatic interactions through its ARM.
Keywords: HIV-1 TAR; Tat; crystal structure; super elongation complex; transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

