Viral RNA structure-based strategies to manipulate translation
- PMID: 30514982
- PMCID: PMC6452865
- DOI: 10.1038/s41579-018-0117-x
Viral RNA structure-based strategies to manipulate translation
Abstract
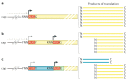
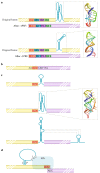
Viruses must co-opt the cellular translation machinery to produce progeny virions. Eukaryotic viruses have evolved a variety of ways to manipulate the cellular translation apparatus, in many cases using elegant RNA-centred strategies. Viral RNAs can alter or control every phase of protein synthesis and have diverse targets, mechanisms and structures. In addition, as cells attempt to limit infection by downregulating translation, some of these viral RNAs enable the virus to overcome this response or even take advantage of it to promote viral translation over cellular translation. In this Review, we present important examples of viral RNA-based strategies to exploit the cellular translation machinery. We describe what is understood of the structures and mechanisms of diverse viral RNA elements that alter or regulate translation, the advantages that are conferred to the virus and some of the major unknowns that provide motivation for further exploration.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

