DockerBIO: web application for efficient use of bioinformatics Docker images
- PMID: 30515360
- PMCID: PMC6266945
- DOI: 10.7717/peerj.5954
DockerBIO: web application for efficient use of bioinformatics Docker images
Abstract
Background and objective: Docker is a light containerization program that shows almost the same performance as a local environment. Recently, many bioinformatics tools have been distributed as Docker images that include complex settings such as libraries, configurations, and data if needed, as well as the actual tools. Users can simply download and run them without making the effort to compile and configure them, and can obtain reproducible results. In spite of these advantages, several problems remain. First, there is a lack of clear standards for distribution of Docker images, and the Docker Hub often provides multiple images with the same objective but different uses. For these reasons, it can be difficult for users to learn how to select and use them. Second, Docker images are often not suitable as a component of a pipeline, because many of them include big data. Moreover, a group of users can have difficulties when sharing a pipeline composed of Docker images. Users of a group may modify scripts or use different versions of the data, which causes inconsistent results.
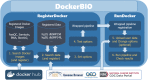
Methods and results: To handle the problems described above, we developed a Java web application, DockerBIO, which provides reliable, verified, light-weight Docker images for various bioinformatics tools and for various kinds of reference data. With DockerBIO, users can easily build a pipeline with tools and data registered at DockerBIO, and if necessary, users can easily register new tools or data. Built pipelines are registered in DockerBIO, which provides an efficient running environment for the pipelines registered at DockerBIO. This enables user groups to run their pipelines without expending much effort to copy and modify them.
Keywords: Bioinformatics; DNA pipeline; DNA-Seq; Docker; Dockerbio; Mygenomebox; NGS pipeline; RNA pipeline; RNA-Seq.
Conflict of interest statement
ChangHyuk Kwon and Jason Kim are employed by MyGenomeBox, Co.
Figures
Similar articles
-
BioShaDock: a community driven bioinformatics shared Docker-based tools registry.F1000Res. 2015 Dec 14;4:1443. doi: 10.12688/f1000research.7536.1. eCollection 2015. F1000Res. 2015. PMID: 26913191 Free PMC article.
-
An open RNA-Seq data analysis pipeline tutorial with an example of reprocessing data from a recent Zika virus study.F1000Res. 2016 Jul 5;5:1574. doi: 10.12688/f1000research.9110.1. eCollection 2016. F1000Res. 2016. PMID: 27583132 Free PMC article.
-
Reproducible bioinformatics project: a community for reproducible bioinformatics analysis pipelines.BMC Bioinformatics. 2018 Oct 15;19(Suppl 10):349. doi: 10.1186/s12859-018-2296-x. BMC Bioinformatics. 2018. PMID: 30367595 Free PMC article.
-
BGDMdocker: a Docker workflow for data mining and visualization of bacterial pan-genomes and biosynthetic gene clusters.PeerJ. 2017 Nov 30;5:e3948. doi: 10.7717/peerj.3948. eCollection 2017. PeerJ. 2017. PMID: 29204317 Free PMC article.
-
CREDO: a friendly Customizable, REproducible, DOcker file generator for bioinformatics applications.BMC Bioinformatics. 2024 Mar 12;25(1):110. doi: 10.1186/s12859-024-05695-9. BMC Bioinformatics. 2024. PMID: 38475691 Free PMC article.
Cited by
-
Democratizing bioinformatics through easily accessible software platforms for non-experts in the field.Biotechniques. 2022 Feb;72(2):36-38. doi: 10.2144/btn-2021-0060. Epub 2022 Jan 21. Biotechniques. 2022. PMID: 35060754 Free PMC article. No abstract available.
References
-
- Andrews S. Babraham bioinformatics—FastQC a quality control tool for high throughput sequence data. 2015. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Da Veiga Leprevost F, Grüning BA, Alves Aflitos S, Röst HL, Uszkoreit J, Barsnes H, Vaudel M, Moreno P, Gatto L, Weber J, Bai M, Jimenez RC, Sachsenberg T, Pfeuffer J, Vera Alvarez R, Griss J, Nesvizhskii AI, Perez-Riverol Y. BioContainers: an open-source and community-driven framework for software standardization. Bioinformatics. 2017;33(16):2580–2582. doi: 10.1093/bioinformatics/btx192. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

