Development and Application of FASA, a Model for Quantifying Fatty Acid Metabolism Using Stable Isotope Labeling
- PMID: 30517876
- PMCID: PMC6432944
- DOI: 10.1016/j.celrep.2018.11.041
Development and Application of FASA, a Model for Quantifying Fatty Acid Metabolism Using Stable Isotope Labeling
Abstract
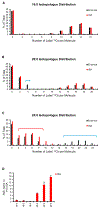
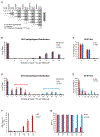
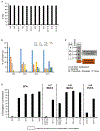
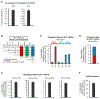
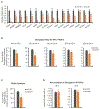
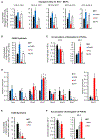
It is well understood that fatty acids can be synthesized, imported, and modified to meet requisite demands in cells. However, following the movement of fatty acids through the multiplicity of these metabolic steps has remained difficult. To better address this problem, we developed Fatty Acid Source Analysis (FASA), a model that defines the contribution of synthesis, import, and elongation pathways to fatty acid homeostasis in saturated, monounsaturated, and polyunsaturated fatty acid pools. Application of FASA demonstrated that elongation can be a major contributor to cellular fatty acid content and showed that distinct pro-inflammatory stimuli (e.g., Toll-like receptors 2, 3, or 4) specifically reprogram homeostasis of fatty acids by differential utilization of synthetic and elongation pathways in macrophages. In sum, this modeling approach significantly advances our ability to interrogate cellular fatty acid metabolism and provides insight into how cells dynamically reshape their lipidomes in response to metabolic or inflammatory signals.
Keywords: fatty acid homeostasis; fatty acid modeling; stable isotope labeling.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Bulusu V, Tumanov S, Michalopoulou E, van den Broek NJ, MacKay G, Nixon C, Dhayade S, Schug ZT, Vande Voorde J, Blyth K, et al. (2017). Acetate Recapturing by Nuclear Acetyl-CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Rep. 18, 647–658. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

