IMPDH2 promotes colorectal cancer progression through activation of the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 signaling pathways
- PMID: 30518405
- PMCID: PMC6282329
- DOI: 10.1186/s13046-018-0980-3
IMPDH2 promotes colorectal cancer progression through activation of the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 signaling pathways
Abstract
Background: Inosine 5'-monophosphate dehydrogenase type II (IMPDH2) was originally identified as an oncogene in several human cancers. However, the clinical significance and biological role of IMPDH2 remain poorly understood in colorectal cancer (CRC).
Methods: Quantitative real-time polymerase chain reaction (qPCR), western blotting analysis, the Cancer Genome Atlas (TCGA) data mining and immunohistochemistry were employed to examine IMPDH2 expression in CRC cell lines and tissues. A series of in-vivo and in-vitro assays were performed to demonstrate the function of IMPDH2 and its possible mechanisms in CRC.
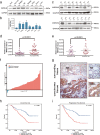
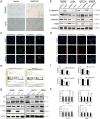
Results: IMPDH2 was upregulated in CRC cells and tissues at both mRNA and protein level. High IMPDH2 expression was closely associated with T stage, lymph node state, distant metastasis, lymphovascular invasion and clinical stage, and significantly correlated with poor survival of CRC patients. Further study revealed that overexpression of IMPDH2 significantly promoted the proliferation, invasion, migration and epithelial-mesenchymal transition (EMT) of CRC cells in vitro and accelerated xenograft tumour growth in nude mice. On the contrary, knockdown of IMPDH2 achieved the opposite effect. Gene set enrichment analysis (GSEA) showed that the gene set related to cell cycle was linked to upregulation of IMPDH2 expression. Our study verified that overexpressing IMPDH2 could promote G1/S phase cell cycle transition through activation of PI3K/AKT/mTOR and PI3K/AKT/FOXO1 pathways and facilitate cell invasion, migration and EMT by regulating PI3K/AKT/mTOR pathway.
Conclusions: These results suggest that IMPDH2 plays an important role in the development and progression of human CRC and may serve as a novel prognostic biomarker and therapeutic target for CRC.
Keywords: Cell cycle; Colorectal cancer; EMT; IMPDH2; Proliferation.
Conflict of interest statement
Ethics approval and consent to participate
This study was approved by the Ethics Committees of Nanfang Hospital, Southern Medical University. Written informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Nagai M, Natsumeda Y, Konno Y, Hoffman R, Irino S, Weber G. Selective up-regulation of type II inosine 5′-monophosphate dehydrogenase messenger RNA expression in human leukemias. Cancer Res. 1991;51:3886–3890. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

