Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry
- PMID: 30518681
- PMCID: PMC6328018
- DOI: 10.1172/jci.insight.124928
Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry
Abstract
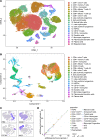
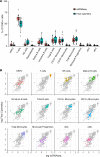
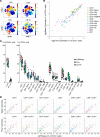
New techniques for single-cell analysis have led to insights into hematopoiesis and the immune system, but the ability of these techniques to cross-validate and reproducibly identify the biological variation in diverse human samples is currently unproven. We therefore performed a comprehensive assessment of human bone marrow cells using both single-cell RNA sequencing and multiparameter flow cytometry from 20 healthy adult human donors across a broad age range. These data characterize variation between healthy donors as well as age-associated changes in cell population frequencies. Direct comparison of techniques revealed discrepancy in the quantification of T lymphocyte and natural killer cell populations. Orthogonal validation of immunophenotyping using mass cytometry demonstrated a strong correlation with flow cytometry. Technical replicates using single-cell RNA sequencing matched robustly, while biological replicates showed variation. Given the increasing use of single-cell technologies in translational research, this resource serves as an important reference data set and highlights opportunities for further refinement.
Keywords: Adaptive immunity; Bone marrow; Bone marrow differentiation; Hematology; Immunology.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

