IL-6 mediates platinum-induced enrichment of ovarian cancer stem cells
- PMID: 30518684
- PMCID: PMC6328027
- DOI: 10.1172/jci.insight.122360
IL-6 mediates platinum-induced enrichment of ovarian cancer stem cells
Abstract
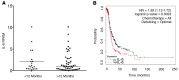
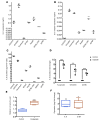
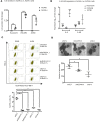
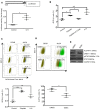
In high-grade serous ovarian cancer (OC), chemotherapy eliminates the majority of tumor cells, leaving behind residual tumors enriched in OC stem cells (OCSC). OCSC, defined as aldehyde dehydrogenase-positive (ALDH+), persist and contribute to tumor relapse. Inflammatory cytokine IL-6 is elevated in residual tumors after platinum treatment, and we hypothesized that IL-6 plays a critical role in platinum-induced OCSC enrichment. We demonstrate that IL-6 regulates stemness features of OCSC driven by ALDH1A1 expression and activity. We show that platinum induces IL-6 secretion by cancer-associated fibroblasts in the tumor microenvironment, promoting OCSC enrichment in residual tumors after chemotherapy. By activating STAT3 and upregulating ALDH1A1 expression, IL-6 treatment converted non-OCSC to OCSC. Having previously shown altered DNA methylation in OCSC, we show here that IL-6 induces DNA methyltransferase 1 (DNMT1) expression and the hypomethylating agent (HMA) guadecitabine induced differentiation of OCSC and reduced - but did not completely eradicate - OCSC. IL-6 neutralizing antibody (IL-6-Nab) combined with HMA fully eradicated OCSC, and the combination blocked IL-6/IL6-R/pSTAT3-mediated ALDH1A1 expression and eliminated OCSC in residual tumors that persisted in vivo after chemotherapy. We conclude that IL-6 signaling blockade combined with an HMA can eliminate OCSC after platinum treatment, supporting this strategy to prevent tumor recurrence after standard chemotherapy.
Keywords: Cancer; Epigenetics; Immunotherapy; Inflammation; Stem cells.
Conflict of interest statement
Figures


References
-
- Berek JS, et al. Advanced epithelial ovarian cancer: 1998 consensus statements. Ann Oncol. 1999;10 Suppl 1:87–92. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

