A two-step strategy for identification of plasma protein biomarkers for endometrial and ovarian cancer
- PMID: 30519148
- PMCID: PMC6271635
- DOI: 10.1186/s12014-018-9216-y
A two-step strategy for identification of plasma protein biomarkers for endometrial and ovarian cancer
Abstract
Background: Over 500,000 women worldwide are diagnosed with ovarian or endometrial cancer each year. We have used a two-step strategy to identify plasma proteins that could be used to improve the diagnosis of women with an indication of gynecologic tumor and in population screening.
Methods: In the discovery step we screened 441 proteins in plasma using the proximity extension assay (PEA) and five Olink Multiplex assays (CVD II, CVD III, INF I, ONC II, NEU I) in women with ovarian cancer (n = 106), endometrial cancer (n = 74), benign ovarian tumors (n = 150) and healthy population controls (n = 399). Based on the discovery analyses a set of 27 proteins were selected and two focused multiplex PEA assays were developed. In a replication step the focused assays were used to study an independent set of cases with ovarian cancer (n = 280), endometrial cancer (n = 228), women with benign ovarian tumors (n = 76) and healthy controls (n = 57).
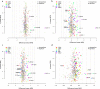
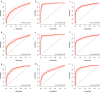
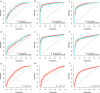
Results: In the discovery step, 27 proteins that showed an association to cancer status were identified. In the replication analyses, the focused assays distinguished benign tumors from ovarian cancer stage III-IV with a sensitivity of 0.88 and specificity of 0.92 (AUC = 0.92). The assays had a significantly higher AUC for distinguishing benign tumors from late stage ovarian cancer than using CA125 and HE4 (p = 9.56e-22). Also, population controls could be distinguished from ovarian cancer stage III-IV with a sensitivity of 0.85 and a specificity of 0.92 (AUC = 0.89).
Conclusion: The PEA assays represent useful tools for identification of new biomarkers for gynecologic cancers. The selected protein assays could be used to distinguish benign tumors from ovarian and endometrial cancer in women diagnosed with an unknown suspicious pelvic mass. The panels could also be used in population screening, for identification of women in need of specialized gynecologic transvaginal ultrasound examination.
Funding: The Swedish Cancer Foundation, Vinnova (SWELIFE), The Foundation for Strategic Research (SSF), Assar Gabrielsson Foundation.
Keywords: Diagnostics; Endometrial cancer; Ovarian cancer; Proximity extension assay (PEA); Sensitivity; Specificity.
Figures




Similar articles
-
A multiplex biomarker assay improves the diagnostic performance of HE4 and CA125 in ovarian tumor patients.PLoS One. 2020 Oct 19;15(10):e0240418. doi: 10.1371/journal.pone.0240418. eCollection 2020. PLoS One. 2020. PMID: 33075095 Free PMC article.
-
A multiplex platform for the identification of ovarian cancer biomarkers.Clin Proteomics. 2017 Oct 10;14:34. doi: 10.1186/s12014-017-9169-6. eCollection 2017. Clin Proteomics. 2017. PMID: 29051715 Free PMC article.
-
[The values of serum human epididymis secretory protein 4 and CA(125) assay in the diagnosis of ovarian malignancy].Zhonghua Fu Chan Ke Za Zhi. 2008 Dec;43(12):931-6. Zhonghua Fu Chan Ke Za Zhi. 2008. PMID: 19134334 Chinese.
-
The emerging role of HE4 in the evaluation of epithelial ovarian and endometrial carcinomas.Oncology (Williston Park). 2013 Jun;27(6):548-56. Oncology (Williston Park). 2013. PMID: 23909069 Free PMC article. Review.
-
Clinical statistics of gynecologic cancers in Japan.J Gynecol Oncol. 2017 Mar;28(2):e32. doi: 10.3802/jgo.2017.28.e32. Epub 2017 Feb 10. J Gynecol Oncol. 2017. PMID: 28198168 Free PMC article. Review.
Cited by
-
Unlocking the potential of tumor-derived DNA in urine for cancer detection: methodological challenges and opportunities.Mol Oncol. 2025 Jul;19(7):1918-1934. doi: 10.1002/1878-0261.13628. Epub 2024 Mar 10. Mol Oncol. 2025. PMID: 38462745 Free PMC article. Review.
-
Significance of Midkine Signaling in Women's Cancers: Novel Biomarker and Therapeutic Target.Int J Mol Sci. 2025 May 17;26(10):4809. doi: 10.3390/ijms26104809. Int J Mol Sci. 2025. PMID: 40429950 Free PMC article. Review.
-
Multi-Steroid Profiling and Machine Learning Reveal Androgens as Candidate Biomarkers for Endometrial Cancer Diagnosis: A Case-Control Study.Cancers (Basel). 2025 May 16;17(10):1679. doi: 10.3390/cancers17101679. Cancers (Basel). 2025. PMID: 40427176 Free PMC article.
-
Proteomics technologies for cancer liquid biopsies.Mol Cancer. 2022 Feb 15;21(1):53. doi: 10.1186/s12943-022-01526-8. Mol Cancer. 2022. PMID: 35168611 Free PMC article. Review.
-
High throughput proteomics identifies a high-accuracy 11 plasma protein biomarker signature for ovarian cancer.Commun Biol. 2019 Jun 20;2:221. doi: 10.1038/s42003-019-0464-9. eCollection 2019. Commun Biol. 2019. PMID: 31240259 Free PMC article.
References
-
- Ferley, GLOBOSCAN 2012: Estimated Cancer incidence, mortality and prevlance worldwide 2012. 2012.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
