Identification, Expression, and Functional Analysis of the Group IId WRKY Subfamily in Upland Cotton (Gossypium hirsutum L.)
- PMID: 30519251
- PMCID: PMC6259137
- DOI: 10.3389/fpls.2018.01684
Identification, Expression, and Functional Analysis of the Group IId WRKY Subfamily in Upland Cotton (Gossypium hirsutum L.)
Abstract
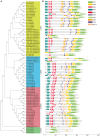
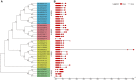
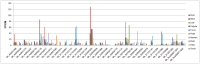
WRKY transcription factors have diverse functions in regulating stress response, leaf senescence, and plant growth and development. However, knowledge of the group IId WRKY subfamily in cotton is largely absent. This study identified 34 group IId WRKY genes in the Gossypium hirsutum genome, and their genomic loci were investigated. Members clustered together in the phylogenetic tree had similar motif compositions and gene structural features, revealing similarity and conservation within group IId WRKY genes. During the evolutionary process, 14 duplicated genes appeared to undergo purification selection. Public RNA-seq data were used to examine the expression patterns of group IId WRKY genes in various tissues and under drought and salt stress conditions. Ten highly expressed genes were identified, and the ten candidate genes revealed distinct expression patterns under drought and salt treatments by qRT-PCR analysis. Among them, Gh_A11G1801 was used for functional characterization. GUS activity was differentially induced by various stresses in Gh_A11G1801p::GUS transgenic Arabidopsis plants. The virus-induced gene silencing (VIGS) of Gh_A11G1801 resulted in drought sensitivity in cotton plants, which was accompanied by elevated malondialdehyde (MDA) content and reduced catalase (CAT) content. Taken together, these findings obtained in this study provide valuable resources for further studying group IId WRKY genes in cotton. Our results also enrich the gene resources for the genetic improvements of cotton varieties that are suitable for growth in stressful conditions.
Keywords: Gh_A11G1801; expression patterns; group IId WRKY genes; stress response; upland cotton.
Figures


Similar articles
-
Characterization and functional analysis of GhWRKY42, a group IId WRKY gene, in upland cotton (Gossypium hirsutum L.).BMC Genet. 2018 Jul 30;19(1):48. doi: 10.1186/s12863-018-0653-4. BMC Genet. 2018. PMID: 30060731 Free PMC article.
-
Identification of the Group III WRKY Subfamily and the Functional Analysis of GhWRKY53 in Gossypium hirsutum L.Plants (Basel). 2021 Jun 17;10(6):1235. doi: 10.3390/plants10061235. Plants (Basel). 2021. PMID: 34204463 Free PMC article.
-
Functional characterization of Gh_A08G1120 (GH3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton.BMC Genet. 2019 Jul 23;20(1):62. doi: 10.1186/s12863-019-0756-6. BMC Genet. 2019. PMID: 31337336 Free PMC article.
-
The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production.Plant Cell Physiol. 2014 Dec;55(12):2060-76. doi: 10.1093/pcp/pcu133. Epub 2014 Sep 26. Plant Cell Physiol. 2014. PMID: 25261532
-
Role of WRKY Transcription Factors in Regulation of Abiotic Stress Responses in Cotton.Life (Basel). 2022 Sep 9;12(9):1410. doi: 10.3390/life12091410. Life (Basel). 2022. PMID: 36143446 Free PMC article. Review.
Cited by
-
Transcription Factor GarWRKY5 Is Involved in Salt Stress Response in Diploid Cotton Species (Gossypium aridum L.).Int J Mol Sci. 2019 Oct 23;20(21):5244. doi: 10.3390/ijms20215244. Int J Mol Sci. 2019. PMID: 31652670 Free PMC article.
-
Identification of SNAT Family Genes Suggests GhSNAT3D Functional Reponse to Melatonin Synthesis Under Salinity Stress in Cotton.Front Mol Biosci. 2022 Feb 10;9:843814. doi: 10.3389/fmolb.2022.843814. eCollection 2022. Front Mol Biosci. 2022. PMID: 35223998 Free PMC article.
-
Transcriptomic Analysis of Short-Term Salt Stress Response in Watermelon Seedlings.Int J Mol Sci. 2020 Aug 21;21(17):6036. doi: 10.3390/ijms21176036. Int J Mol Sci. 2020. PMID: 32839408 Free PMC article.
-
Comprehensive Genome-Wide Analysis of Thaumatin-Like Gene Family in Four Cotton Species and Functional Identification of GhTLP19 Involved in Regulating Tolerance to Verticillium dahlia and Drought.Front Plant Sci. 2020 Oct 20;11:575015. doi: 10.3389/fpls.2020.575015. eCollection 2020. Front Plant Sci. 2020. PMID: 33193513 Free PMC article.
-
Selection of stable reference genes for accurate reverse-transcription quantitative PCR in cotton-herbivore studies using virus-induced gene silencing.Sci Rep. 2025 Jul 8;15(1):24482. doi: 10.1038/s41598-025-08940-0. Sci Rep. 2025. PMID: 40628827 Free PMC article.
References
-
- Cai C., Niu E., Du H., Zhao L., Feng Y., Guo W. (2014a). Genome-wide analysis of the WRKY transcription factor gene family in Gossypium raimondii and the expression of orthologs in cultivated tetraploid cotton. Crop J. 2, 87–101. 10.1016/j.cj.2014.03.001 - DOI
-
- Cai R. H., Zhao Y., Wang Y. F., Lin Y. X., Peng X. J., Li Q., et al. (2014b). Overexpression of a maize WRKY58 gene enhances drought and salt tolerance in transgenic rice. Plant Cell Tiss. Organ Cult. 119, 565–577. 10.1007/s11240-014-0556-7 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

