The transcription factors TFE3 and TFEB amplify p53 dependent transcriptional programs in response to DNA damage
- PMID: 30520728
- PMCID: PMC6292694
- DOI: 10.7554/eLife.40856
The transcription factors TFE3 and TFEB amplify p53 dependent transcriptional programs in response to DNA damage
Abstract
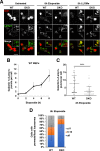
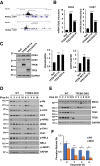
The transcription factors TFE3 and TFEB cooperate to regulate autophagy induction and lysosome biogenesis in response to starvation. Here we demonstrate that DNA damage activates TFE3 and TFEB in a p53 and mTORC1 dependent manner. RNA-Seq analysis of TFEB/TFE3 double-knockout cells exposed to etoposide reveals a profound dysregulation of the DNA damage response, including upstream regulators and downstream p53 targets. TFE3 and TFEB contribute to sustain p53-dependent response by stabilizing p53 protein levels. In TFEB/TFE3 DKOs, p53 half-life is significantly decreased due to elevated Mdm2 levels. Transcriptional profiles of genes involved in lysosome membrane permeabilization and cell death pathways are dysregulated in TFEB/TFE3-depleted cells. Consequently, prolonged DNA damage results in impaired LMP and apoptosis induction. Finally, expression of multiple genes implicated in cell cycle control is altered in TFEB/TFE3 DKOs, revealing a previously unrecognized role of TFEB and TFE3 in the regulation of cell cycle checkpoints in response to stress.
Keywords: DNA Damage; TFE3; TFEB; cell biology; cell cycle; lysosomes; mouse; p53.
Conflict of interest statement
EJ, OB, JM, MP, IT, RP No competing interests declared
Figures


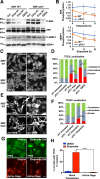


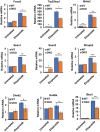
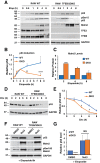
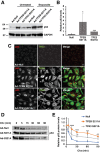






References
-
- Blessing AM, Rajapakshe K, Reddy Bollu L, Shi Y, White MA, Pham AH, Lin C, Jonsson P, Cortes CJ, Cheung E, La Spada AR, Bast RC, Merchant FA, Coarfa C, Frigo DE. Transcriptional regulation of core autophagy and lysosomal genes by the androgen receptor promotes prostate cancer progression. Autophagy. 2017;13:506–521. doi: 10.1080/15548627.2016.1268300. - DOI - PMC - PubMed
-
- Bretou M, Sáez PJ, Sanséau D, Maurin M, Lankar D, Chabaud M, Spampanato C, Malbec O, Barbier L, Muallem S, Maiuri P, Ballabio A, Helft J, Piel M, Vargas P, Lennon-Duménil AM. Lysosome signaling controls the migration of dendritic cells. Science Immunology. 2017;2:eaak9573. doi: 10.1126/sciimmunol.aak9573. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

