Molecular basis for the increased affinity of an RNA recognition motif with re-engineered specificity: A molecular dynamics and enhanced sampling simulations study
- PMID: 30521520
- PMCID: PMC6307825
- DOI: 10.1371/journal.pcbi.1006642
Molecular basis for the increased affinity of an RNA recognition motif with re-engineered specificity: A molecular dynamics and enhanced sampling simulations study
Abstract
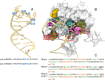
The RNA recognition motif (RRM) is the most common RNA binding domain across eukaryotic proteins. It is therefore of great value to engineer its specificity to target RNAs of arbitrary sequence. This was recently achieved for the RRM in Rbfox protein, where four mutations R118D, E147R, N151S, and E152T were designed to target the precursor to the oncogenic miRNA 21. Here, we used a variety of molecular dynamics-based approaches to predict specific interactions at the binding interface. Overall, we have run approximately 50 microseconds of enhanced sampling and plain molecular dynamics simulations on the engineered complex as well as on the wild-type Rbfox·pre-miRNA 20b from which the mutated systems were designed. Comparison with the available NMR data on the wild type molecules (protein, RNA, and their complex) served to establish the accuracy of the calculations. Free energy calculations suggest that further improvements in affinity and selectivity are achieved by the S151T replacement.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Targeted inhibition of oncogenic miR-21 maturation with designed RNA-binding proteins.Nat Chem Biol. 2016 Sep;12(9):717-23. doi: 10.1038/nchembio.2128. Epub 2016 Jul 18. Nat Chem Biol. 2016. PMID: 27428511 Free PMC article.
-
Exploring the molecular basis of RNA recognition by the dimeric RNA-binding protein via molecular simulation methods.RNA Biol. 2016 Nov;13(11):1133-1143. doi: 10.1080/15476286.2016.1223007. Epub 2016 Sep 3. RNA Biol. 2016. PMID: 27592836 Free PMC article.
-
Integrated structural biology to unravel molecular mechanisms of protein-RNA recognition.Methods. 2017 Apr 15;118-119:119-136. doi: 10.1016/j.ymeth.2017.03.015. Epub 2017 Mar 16. Methods. 2017. PMID: 28315749 Review.
-
The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets.RNA Biol. 2014;11(10):1250-61. doi: 10.1080/15476286.2014.996069. RNA Biol. 2014. PMID: 25584704 Free PMC article.
-
RRM-RNA recognition: NMR or crystallography…and new findings.Curr Opin Struct Biol. 2013 Feb;23(1):100-8. doi: 10.1016/j.sbi.2012.11.006. Epub 2012 Dec 14. Curr Opin Struct Biol. 2013. PMID: 23253355 Review.
Cited by
-
Disclosing the Impact of Carcinogenic SF3b Mutations on Pre-mRNA Recognition Via All-Atom Simulations.Biomolecules. 2019 Oct 21;9(10):633. doi: 10.3390/biom9100633. Biomolecules. 2019. PMID: 31640290 Free PMC article.
-
Spontaneous Membrane Nanodomain Formation in the Absence or Presence of the Neurotransmitter Serotonin.Front Cell Dev Biol. 2020 Nov 30;8:601145. doi: 10.3389/fcell.2020.601145. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33330494 Free PMC article.
-
An overview of structural approaches to study therapeutic RNAs.Front Mol Biosci. 2022 Oct 28;9:1044126. doi: 10.3389/fmolb.2022.1044126. eCollection 2022. Front Mol Biosci. 2022. PMID: 36387283 Free PMC article. Review.
-
RNA-protein complexes and force field polarizability.Front Chem. 2023 Jun 22;11:1217506. doi: 10.3389/fchem.2023.1217506. eCollection 2023. Front Chem. 2023. PMID: 37426330 Free PMC article.
-
In Vitro Evolution Reveals Noncationic Protein-RNA Interaction Mediated by Metal Ions.Mol Biol Evol. 2022 Mar 2;39(3):msac032. doi: 10.1093/molbev/msac032. Mol Biol Evol. 2022. PMID: 35137196 Free PMC article.
References
-
- Burd C, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

