Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology
- PMID: 30521609
- PMCID: PMC6283538
- DOI: 10.1371/journal.pone.0208585
Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology
Abstract
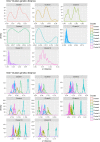
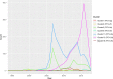
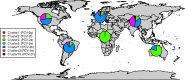
Porcine circovirus 2 (PCV-2) is one of the most widespread viral infections of swine, causing a remarkable economic impact because of direct losses and indirect costs for its control. As other ssDNA viruses, PCV-2 is characterized by a high evolutionary rate, leading to the emergence of a plethora of variants with different biological and epidemiological features. Over time, several attempts have been made to organize PCV-2 genetic heterogeneity in recognized genotypes. This categorization has clearly simplified the epidemiological investigations, allowing to identify different spatial and temporal patterns among genotypes. Additionally, variable virulence and vaccine effectiveness have also been hypothesized. However, the rapid increase in sequencing activity, coupled with the per se high viral variability, has challenged the previously established nomenclature, leading to the definition of several study-specific genotypes and hindering the capability of performing comparable epidemiological studies. Based on these premises, an updated classification scheme is herein reported. Recognizing the impossibility of defining a clear inter-cluster p-distance cut-off, the present study proposes a phylogeny-grounded genotype definition based on three criteria: maximum intra-genotype p-distance of 13% (calculated on the ORF2 gene), bootstrap support at the corresponding internal node higher than 70% and at least 15 available sequences. This scheme allowed defining 8 genotypes (PCV-2a to PCV-2h), which six of those had been previously proposed. To minimize the inconvenience of implementing a new classification, the most common names already adopted have been maintained when possible. The analysis of sequence-associated metadata highlighted a highly unbalanced sequencing activity in terms of geographical, host and temporal distribution. The PCV-2 molecular epidemiology scenario appears therefore characterized by a severe bias that could lead to spurious associations between genetic and epidemiological/biological viral features. While the suggested classification can establish a "common language" for future studies, further efforts should be paid to achieve a more homogeneous and informative representation of the PCV-2 global scenario.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Olvera A, Cortey M, Segalés J. Molecular evolution of porcine circovirus type 2 genomes: Phylogeny and clonality.Virology. 2007;357: 175–185. 10.1016/j.virol.2006.07.047 - DOI - PubMed
-
- Segalés J, Olvera A, Grau-Roma L, Charreyre C, Nauwynck H, Larsen L, et al. PCV-2 genotype definition and nomenclature. Vet Rec. 2008;162: 867–868. 10.1136/vr.162.26.867 - DOI - PubMed
-
- Dupont K, Nielsen EO, Bækbo P, Larsen LE. Genomic analysis of PCV2 isolates from Danish archives and a current PMWS case-control study supports a shift in genotypes with time. Vet Microbiol. 2008;128: 56–64. 10.1016/j.vetmic.2007.09.016 - DOI - PubMed
-
- Franzo G, Cortey M, Castro AMMG de, Piovezan U, Szabo MPJ, Drigo M, et al. Genetic characterisation of Porcine circovirus type 2 (PCV2) strains from feral pigs in the Brazilian Pantanal: An opportunity to reconstruct the history of PCV2 evolution. Vet Microbiol. 2015;178: 158–162. 10.1016/j.vetmic.2015.05.003 - DOI - PubMed
-
- Liu X, Wang FX, Zhu HW, Sun N, Wu H. Phylogenetic analysis of porcine circovirus type 2 (PCV2) isolates from China with high homology to PCV2c. Arch Virol. 2016;161: 1591–1599. 10.1007/s00705-016-2823-x - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

