Viability-based quantification of antibiotic resistance genes and human fecal markers in wastewater effluent and receiving waters
- PMID: 30522032
- PMCID: PMC6526933
- DOI: 10.1016/j.scitotenv.2018.11.325
Viability-based quantification of antibiotic resistance genes and human fecal markers in wastewater effluent and receiving waters
Abstract
Antibiotic resistance is a public health issue with links to environmental sources of antibiotic resistance genes (ARGs). ARGs from nonviable sources may pose a hazard given the potential for transformation whereas ARGs in viable sources may proliferate during host growth or conjugation. In this study, ARGs in the effluent from three municipal wastewater treatment plants (WWTPs) and the receiving surface waters were investigated using a viability-based qPCR technique (vPCR) with propidium monoazide (PMA). ARGs sul1, tet(G), and blaTEM, fecal indicator marker BacHum, and 16S rRNA gene copies/mL were found to be significantly lower in viable-cells than in total concentrations for WWTP effluent. Viable-cell and total gene copy concentrations were similar in downstream samples except for tet(G). Differences with respect to season in the prevalence of nonviable ARGs in surface water or WWTP effluent were not observed. The results of this study indicate that qPCR may overestimate viable-cell ARGs and fecal indicator genes in WWTP effluent but not necessarily in the surface water >1.8 km downstream.
Keywords: Antibiotic resistance genes; Chlorination; Propidium monoazide; Wastewater; vPCR.
Copyright © 2018 Elsevier B.V. All rights reserved.
Figures

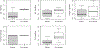

References
-
- Banihashemi A, Van Dyke MI, Huck PM. Long-amplicon propidium monoazide-PCR enumeration assay to detect viable Campylobacter and Salmonella. J Appl Microbiol 2012; 113: 863–73. - PubMed
-
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013 In: Department of Health and Human Services, editor, 2013.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

