The split protein phosphatase system
- PMID: 30523060
- PMCID: PMC6282683
- DOI: 10.1042/BCJ20170726
The split protein phosphatase system
Abstract
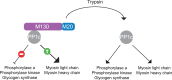
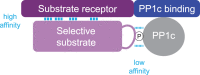
Reversible phosphorylation of proteins is a post-translational modification that regulates all aspect of life through the antagonistic action of kinases and phosphatases. Protein kinases are well characterized, but protein phosphatases have been relatively neglected. Protein phosphatase 1 (PP1) catalyzes the dephosphorylation of a major fraction of phospho-serines and phospho-threonines in cells and thereby controls a broad range of cellular processes. In this review, I will discuss how phosphatases were discovered, how the view that they were unselective emerged and how recent findings have revealed their exquisite selectivity. Unlike kinases, PP1 phosphatases are obligatory heteromers composed of a catalytic subunit bound to one (or two) non-catalytic subunit(s). Based on an in-depth study of two holophosphatases, I propose the following: selective dephosphorylation depends on the assembly of two components, the catalytic subunit and the non-catalytic subunit, which serves as a high-affinity substrate receptor. Because functional complementation of the two modules is required to produce a selective holophosphatase, one can consider that they are split enzymes. The non-catalytic subunit was often referred to as a regulatory subunit, but it is, in fact, an essential component of the holoenzyme. In this model, a phosphatase and its array of mostly orphan substrate receptors constitute the split protein phosphatase system. The set of potentially generalizable principles outlined in this review may facilitate the study of these poorly understood enzymes and the identification of their physiological substrates.
Keywords: biochemical techniques and resources; intracellular signaling; protein phosphatases.
© 2018 The Author(s).
Conflict of interest statement
A.B. is a founder and Chief Scientific Officer of a small company, CamPhos Therapeutics, which develops phosphatase inhibitors.
Figures








References
-
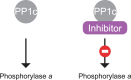
- Brandt H., Killilea S.D. and Lee E.Y.C. (1974) Activation of phosphorylase phosphatase by a novel procedure: evidence for a regulatory mechanism involving the release of a catalytic subunit from enzyme-inhibitor complex(es) of higher molecular weight. Biochem. Biophys. Res. Commun. 61, 598–604 10.1016/0006-291X(74)90999-1 - DOI - PubMed
-
- Cori G.T. and Green A.A. (1943) Crystalline muscle phosphorylase II. Prosthetic group. J. Biol. Chem. 151, 31–38
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

