Molecular High-Grade B-Cell Lymphoma: Defining a Poor-Risk Group That Requires Different Approaches to Therapy
- PMID: 30523719
- PMCID: PMC6338391
- DOI: 10.1200/JCO.18.01314
Molecular High-Grade B-Cell Lymphoma: Defining a Poor-Risk Group That Requires Different Approaches to Therapy
Erratum in
-
Erratum.J Clin Oncol. 2019 Apr 20;37(12):1035. doi: 10.1200/JCO.19.00581. J Clin Oncol. 2019. PMID: 30995418 Free PMC article. No abstract available.
Abstract
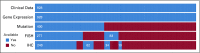
Purpose: Biologic heterogeneity is a feature of diffuse large B-cell lymphoma (DLBCL), and the existence of a subgroup with poor prognosis and phenotypic proximity to Burkitt lymphoma is well known. Conventional cytogenetics identifies some patients with rearrangements of MYC and BCL2 and/or BCL6 (double-hit lymphomas) who are increasingly treated with more intensive chemotherapy, but a more biologically coherent and clinically useful definition of this group is required.
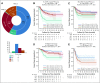
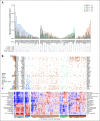
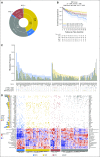
Patients and methods: We defined a molecular high-grade (MHG) group by applying a gene expression-based classifier to 928 patients with DLBCL from a clinical trial that investigated the addition of bortezomib to standard rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) therapy. The prognostic significance of MHG was compared with existing biomarkers. We performed targeted sequencing of 70 genes in 400 patients and explored molecular pathology using gene expression signature databases. Findings were validated in an independent data set.
Results: The MHG group comprised 83 patients (9%), with 75 in the cell-of-origin germinal center B-cell-like group. MYC rearranged and double-hit groups were strongly over-represented in MHG but comprised only one half of the total. Gene expression analysis revealed a proliferative phenotype with a relationship to centroblasts. Progression-free survival rate at 36 months after R-CHOP in the MHG group was 37% (95% CI, 24% to 55%) compared with 72% (95% CI, 68% to 77%) for others, and an analysis of treatment effects suggested a possible positive effect of bortezomib. Double-hit lymphomas lacking the MHG signature showed no evidence of worse outcome than other germinal center B-cell-like cases.
Conclusion: MHG defines a biologically coherent high-grade B-cell lymphoma group with distinct molecular features and clinical outcomes that effectively doubles the size of the poor-prognosis, double-hit group. Patients with MHG may benefit from intensified chemotherapy or novel targeted therapies.
Figures

Comment in
-
Using Gene Expression Profiling to Move Beyond MYC/BCL2 Rearrangements in High-Grade Lymphoma.J Clin Oncol. 2019 Jan 20;37(3):175-177. doi: 10.1200/JCO.18.01910. Epub 2018 Dec 3. J Clin Oncol. 2019. PMID: 30523718 Free PMC article. No abstract available.
References
-
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
-
- Intlekofer AM, Younes A. Precision therapy for lymphoma--current state and future directions. Nat Rev Clin Oncol. 2014;11:585–596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

