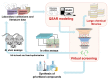
QSAR-Based Virtual Screening: Advances and Applications in Drug Discovery
- PMID: 30524275
- PMCID: PMC6262347
- DOI: 10.3389/fphar.2018.01275
QSAR-Based Virtual Screening: Advances and Applications in Drug Discovery
Abstract
Virtual screening (VS) has emerged in drug discovery as a powerful computational approach to screen large libraries of small molecules for new hits with desired properties that can then be tested experimentally. Similar to other computational approaches, VS intention is not to replace in vitro or in vivo assays, but to speed up the discovery process, to reduce the number of candidates to be tested experimentally, and to rationalize their choice. Moreover, VS has become very popular in pharmaceutical companies and academic organizations due to its time-, cost-, resources-, and labor-saving. Among the VS approaches, quantitative structure-activity relationship (QSAR) analysis is the most powerful method due to its high and fast throughput and good hit rate. As the first preliminary step of a QSAR model development, relevant chemogenomics data are collected from databases and the literature. Then, chemical descriptors are calculated on different levels of representation of molecular structure, ranging from 1D to nD, and then correlated with the biological property using machine learning techniques. Once developed and validated, QSAR models are applied to predict the biological property of novel compounds. Although the experimental testing of computational hits is not an inherent part of QSAR methodology, it is highly desired and should be performed as an ultimate validation of developed models. In this mini-review, we summarize and critically analyze the recent trends of QSAR-based VS in drug discovery and demonstrate successful applications in identifying perspective compounds with desired properties. Moreover, we provide some recommendations about the best practices for QSAR-based VS along with the future perspectives of this approach.
Keywords: cheminformatics; computer-assisted drug design; machine learning; molecular descriptors; virtual screening.
Figures
References
-
- Ban F., Dalal K., Li H., LeBlanc E., Rennie P. S., Cherkasov A. (2017). Best practices of computer-aided drug discovery: lessons learned from the development of a preclinical candidate for prostate cancer with a new mechanism of action. J. Chem. Inf. Model. 57 1018–1028. 10.1021/acs.jcim.7b00137 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials