Specific MHC-I Peptides Are Induced Using PROTACs
- PMID: 30524438
- PMCID: PMC6262898
- DOI: 10.3389/fimmu.2018.02697
Specific MHC-I Peptides Are Induced Using PROTACs
Abstract
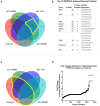
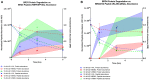
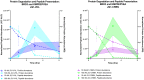
Peptides presented by the class-I major histocompatibility complex (MHC-I) are important targets for immunotherapy. The identification of these peptide targets greatly facilitates the generation of T-cell-based therapeutics. Herein, we report the capability of proteolysis targeting chimera (PROTAC) compounds to induce the presentation of specific MHC class-I peptides derived from endogenous cellular proteins. Using LC-MS/MS, we identified several BET-derived MHC-I peptides induced by treatment with three BET-directed PROTAC compounds. To understand our ability to tune this process, we measured the relative rate of presentation of these peptides under varying treatment conditions using label-free mass spectrometry quantification. We found that the rate of peptide presentation reflected the rate of protein degradation, indicating a direct relationship between PROTAC treatment and peptide presentation. We additionally analyzed the effect of PROTAC treatment on the entire immunopeptidome and found many new peptides that were displayed in a PROTAC-specific fashion: we determined that these identifications map to the BET pathway, as well as, potential off-target or unique-to-PROTAC pathways. This work represents the first evidence of the use of PROTAC compounds to induce the presentation of MHC-I peptides from endogenous cellular proteins, highlighting the capability of PROTAC compounds for the discovery and generation of new targets for immunotherapy.
Keywords: BET; HLA; MHC-I; PROTAC; immunopeptides.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

