TNFR2/BIRC3-TRAF1 signaling pathway as a novel NK cell immune checkpoint in cancer
- PMID: 30524877
- PMCID: PMC6279330
- DOI: 10.1080/2162402X.2017.1386826
TNFR2/BIRC3-TRAF1 signaling pathway as a novel NK cell immune checkpoint in cancer
Abstract
Natural Killer (NK) cells control metastatic dissemination of murine tumors and are an important prognostic factor in several human malignancies. However, tumor cells hijack many of the NK cell functional features compromising their tumoricidal activity. Here, we show a deleterious role of the TNFα/TNFR2/BIRC3/TRAF1 signaling cascade in NK cells from the tumor microenvironment (TME). TNFα induces BIRC3/cIAP2 transcripts and reduces NKp46/NCR1 transcription and surface expression on NK cells, promoting metastases dissemination in mice and poor prognosis in GIST patients. NKp30 engagement, by promoting the release of TNFα, also contributes to BIRC3 upregulation, and more so in patients expressing predominantly NKp30C isoforms. These findings reveal that in the absence of IL-12 or a Th1-geared TME, TNFα can be considered as a negative regulatory cytokine for innate effectors.
Keywords: BIRC3; NK cells; TNFα; TRAF1; cancer; immune checkpoint; immunity.
Figures
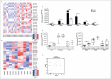
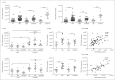
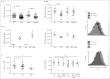



References
Publication types
LinkOut - more resources
Full Text Sources
