Profiling targetable immune checkpoints in osteosarcoma
- PMID: 30524885
- PMCID: PMC6279416
- DOI: 10.1080/2162402X.2018.1475873
Profiling targetable immune checkpoints in osteosarcoma
Abstract
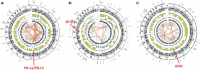
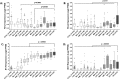
Osteosarcomas are aggressive bone tumors for which therapeutic advances have not improved over several decades. Unlike most pediatric tumors, the osteosarcoma genome is remarkably unstable, characterized by numerous copy number alterations and chromosomal structural aberrations. In this study, we asked if the targetable immune checkpoints CD274 (PD-L1), PDCD1LG2 (PD-L2), CD276 (B7-H3) and IDO1 are impacted by copy number alterations in osteosarcoma. Of the 215 osteosarcoma samples investigated, PD-L1/PD-L2, B7-H3 and IDO1 were independently gained at frequencies of approximately 8-9%, with a cumulative frequency of approximately 24%. RNA sequencing data from two independent cohorts revealed that B7-H3 is the most highly expressed immune checkpoint gene among the four investigated. We also show that IDO1 is preferentially expressed in pediatric solid tumors and that increased protein expression of B7-H3 and IDO1 are significantly associated with inferior survival in patient samples. Using human osteosarcoma cell lines, we demonstrate that IDO1 is gained in MG63 and G292 cells and that the IDO1 inhibitor, epacadostat, inhibits the enzymatic activity of IDO1 in a dose-dependent manner in these cells. Together, these data reveal the genomic and transcriptomic profiles of PD-L1, PD-L2, B7-H3 and IDO1 in osteosarcoma and identifies a potential context for targeted immunotherapeutic intervention in a subset of patients.
Keywords: B7-H3; CD276; IDO1; Osteosarcoma; PD-L1; PD-L2; checkpoint.
Figures







References
-
- Lorenz S, Barøy T, Sun J, Nome T, Vodák D, Bryne JC, Håkelien AM, Fernandez-Cuesta L, Möhlendick B, Rieder H, et al. Unscrambling the genomic chaos of osteosarcoma reveals extensive transcript fusion, recurrent rearrangements and frequent novel TP53 aberrations. Oncotarget. 2016. 7(5):5273–5288. doi:10.18632/oncotarget.6567. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
