Upregulation of tryptophanyl-tRNA synthethase adapts human cancer cells to nutritional stress caused by tryptophan degradation
- PMID: 30524887
- PMCID: PMC6279332
- DOI: 10.1080/2162402X.2018.1486353
Upregulation of tryptophanyl-tRNA synthethase adapts human cancer cells to nutritional stress caused by tryptophan degradation
Abstract
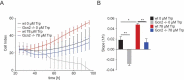
Tryptophan (Trp) metabolism is an important target in immuno-oncology as it represents a powerful immunosuppressive mechanism hijacked by tumors for protection against immune destruction. However, it remains unclear how tumor cells can proliferate while degrading the essential amino acid Trp. Trp is incorporated into proteins after it is attached to its tRNA by tryptophanyl-tRNA synthestases. As the tryptophanyl-tRNA synthestases compete for Trp with the Trp-catabolizing enzymes, the balance between these enzymes will determine whether Trp is used for protein synthesis or is degraded. In human cancers expression of the Trp-degrading enzymes indoleamine-2,3-dioxygenase-1 (IDO1) and tryptophan-2,3-dioxygenase (TDO2) was positively associated with the expression of the tryptophanyl-tRNA synthestase WARS. One mechanism underlying the association between IDO1 and WARS identified in this study is their joint induction by IFNγ released from tumor-infiltrating T cells. Moreover, we show here that IDO1- and TDO2-mediated Trp deprivation upregulates WARS expression by activating the general control non-derepressible-2 (GCN2) kinase, leading to phosphorylation of the eukaryotic translation initiation factor 2α (eIF2α) and induction of activating transcription factor 4 (ATF4). Trp deprivation induced cytoplasmic WARS expression but did not increase nuclear or extracellular WARS levels. GCN2 protected the cells against the effects of Trp starvation and enabled them to quickly make use of Trp for proliferation once it was replenished. Computational modeling of Trp metabolism revealed that Trp deficiency shifted Trp flux towards WARS and protein synthesis. Our data therefore suggest that the upregulation of WARS via IFNγ and/or GCN2-peIF2α-ATF4 signaling protects Trp-degrading cancer cells from excessive intracellular Trp depletion.
Keywords: 3-dioxygenase; Indoleamine-2, 3-dioxygenase; cancer metabolism; immunosuppression; immunosurveillance; inflammation and cancer; nutrients; proliferation; starvation; tRNA synthetase; tryptophan; tryptophan-2; tumor.
Figures





References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
