Single Cell Gene Expression to Understand the Dynamic Architecture of the Heart
- PMID: 30525044
- PMCID: PMC6258739
- DOI: 10.3389/fcvm.2018.00167
Single Cell Gene Expression to Understand the Dynamic Architecture of the Heart
Abstract
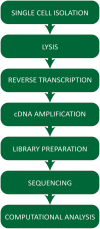
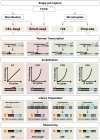
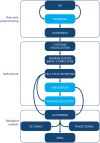
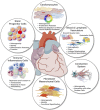
The recent development of single cell gene expression technologies, and especially single cell transcriptomics, have revolutionized the way biologists and clinicians investigate organs and organisms, allowing an unprecedented level of resolution to the description of cell demographics in both healthy and diseased states. Single cell transcriptomics provide information on prevalence, heterogeneity, and gene co-expression at the individual cell level. This enables a cell-centric outlook to define intracellular gene regulatory networks and to bridge toward the definition of intercellular pathways otherwise masked in bulk analysis. The technologies have developed at a fast pace producing a multitude of different approaches, with several alternatives to choose from at any step, including single cell isolation and capturing, lysis, RNA reverse transcription and cDNA amplification, library preparation, sequencing, and computational analyses. Here, we provide guidelines for the experimental design of single cell RNA sequencing experiments, exploring the current options for the crucial steps. Furthermore, we provide a complete overview of the typical data analysis workflow, from handling the raw sequencing data to making biological inferences. Significantly, advancements in single cell transcriptomics have already contributed to outstanding exploratory and functional studies of cardiac development and disease models, as summarized in this review. In conclusion, we discuss achievable outcomes of single cell transcriptomics' applications in addressing unanswered questions and influencing future cardiac clinical applications.
Keywords: RNA-seq; cellular landscape; gene expression; heart; qRT-PCR; single cell; transcriptomics.
Figures