Architectural Dynamics of CaMKII-Actin Networks
- PMID: 30527447
- PMCID: PMC6341221
- DOI: 10.1016/j.bpj.2018.11.006
Architectural Dynamics of CaMKII-Actin Networks
Abstract
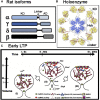
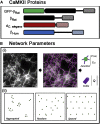
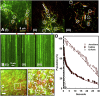
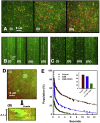
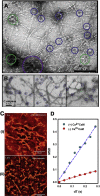
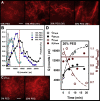
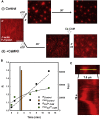
Calcium-calmodulin-dependent kinase II (CaMKII) has an important role in dendritic spine remodeling upon synaptic stimulation. Using fluorescence video microscopy and image analysis, we investigated the architectural dynamics of rhodamine-phalloidin stabilized filamentous actin (F-actin) networks cross-linked by CaMKII. We used automated image analysis to identify F-actin bundles and crossover junctions and developed a dimensionless metric to characterize network architecture. Similar networks were formed by three different CaMKII species with a 10-fold length difference in the linker region between the kinase domain and holoenzyme hub, implying linker length is not a primary determinant of F-actin cross-linking. Electron micrographs showed that at physiological molar ratios, single CaMKII holoenzymes cross-linked multiple F-actin filaments at random, whereas at higher CaMKII/F-actin ratios, filaments bundled. Light microscopy established that the random network architecture resisted macromolecular crowding with polyethylene glycol and blocked ATP-powered compaction by myosin-II miniature filaments. Importantly, the networks disassembled after the addition of calcium-calmodulin and were then spaced within 3 min into compacted foci by myosin motors or more slowly (30 min) aggregated by crowding. Single-molecule total internal reflection fluorescence microscopy showed CaMKII dissociation from surface-immobilized globular actin exhibited a monoexponential dwell-time distribution, whereas CaMKII bound to F-actin networks had a long-lived fraction, trapped at crossover junctions. Release of CaMKII from F-actin, triggered by calcium-calmodulin, was too rapid to measure with flow-cell exchange (<20 s). The residual bound fraction was reduced substantially upon addition of an N-methyl-D-aspartate receptor peptide analog but not ATP. These results provide mechanistic insights to CaMKII-actin interactions at the collective network and single-molecule level. Our findings argue that CaMKII-actin networks in dendritic spines maintain spine size against physical stress. Upon synaptic stimulation, CaMKII is disengaged by calcium-calmodulin, triggering network disassembly, expansion, and subsequent compaction by myosin motors with kinetics compatible with the times recorded for the poststimulus changes in spine volume.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Lisman J., Malenka R.C., Malinow R. Learning mechanisms: the case for CaM-KII. Science. 1997;276:2001–2002. - PubMed
-
- Rongo C., Kaplan J.M. CaMKII regulates the density of central glutamatergic synapses in vivo. Nature. 1999;402:195–199. - PubMed
-
- Herring B.E., Nicoll R.A. Long-term potentiation: from CaMKII to AMPA receptor trafficking. Annu. Rev. Physiol. 2016;78:351–365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

