An interdomain bridge influences RNA binding of the human La protein
- PMID: 30530494
- PMCID: PMC6364776
- DOI: 10.1074/jbc.RA118.003995
An interdomain bridge influences RNA binding of the human La protein
Abstract
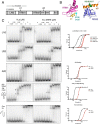
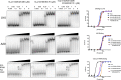
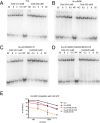
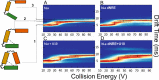
La proteins are RNA chaperones that perform various functions depending on distinct RNA-binding modes and their subcellular localization. In the nucleus, they help process UUU-3'OH-tailed nascent RNA polymerase III transcripts, such as pre-tRNAs, whereas in the cytoplasm they contribute to translation of poly(A)-tailed mRNAs. La accumulation in the nucleus and cytoplasm is controlled by several trafficking elements, including a canonical nuclear localization signal in the extreme C terminus and a nuclear retention element (NRE) in the RNA recognition motif 2 (RRM2) domain. Previous findings indicate that cytoplasmic export of La due to mutation of the NRE can be suppressed by mutations in RRM1, but the mechanism by which the RRM1 and RRM2 domains functionally cooperate is poorly understood. In this work, we use electromobility shift assays (EMSA) to show that mutations in the NRE and RRM1 affect binding of human La to pre-tRNAs but not UUU-3'OH or poly(A) sequences, and we present compensatory mutagenesis data supporting a direct interaction between the RRM1 and RRM2 domains. Moreover, we use collision-induced unfolding and time-resolved hydrogen-deuterium exchange MS analyses to study the conformational dynamics that occur when this interaction is intact or disrupted. Our results suggest that the intracellular distribution of La may be linked to its RNA-binding modes and provide the first evidence for a direct protein-protein interdomain interaction in La proteins.
Keywords: La protein; RNA processing; RNA-binding protein; RNA–protein interaction; SSB; Sjögren syndrome antigen B; intracellular trafficking; mass spectrometry (MS); precursor tRNA (pre-tRNA); protein domain.
© 2019 Marrella et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
-
- Teplova M., Yuan Y.-R., Phan A. T., Malinina L., Ilin S., Teplov A., and Patel D. J. (2006) Structural basis for recognition and sequestration of UUU(OH) 3′ termini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol. Cell 21, 75–85 10.1016/j.molcel.2005.10.027 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

