AcrR and Rex Control Mannitol and Sorbitol Utilization through Their Cross-Regulation of Aldehyde-Alcohol Dehydrogenase (AdhE) in Lactobacillus plantarum
- PMID: 30530710
- PMCID: PMC6365815
- DOI: 10.1128/AEM.02035-18
AcrR and Rex Control Mannitol and Sorbitol Utilization through Their Cross-Regulation of Aldehyde-Alcohol Dehydrogenase (AdhE) in Lactobacillus plantarum
Abstract
Lactobacillus plantarum is a versatile bacterium that occupies a wide range of environmental niches. In this study, we found that a bifunctional aldehyde-alcohol dehydrogenase-encoding gene, adhE, was responsible for L. plantarum being able to utilize mannitol and sorbitol through cross-regulation by two DNA-binding regulators. In L. plantarum NF92, adhE was greatly induced, and the growth of an adhE-disrupted (ΔadhE) strain was repressed when sorbitol or mannitol instead of glucose was used as a carbon source. The results of enzyme activity and metabolite assays demonstrated that AdhE could catalyze the synthesis of ethanol in L. plantarum NF92 when sorbitol or mannitol was used as the carbon source. AcrR and Rex were two transcriptional factors screened by an affinity isolation method and verified to regulate the expression of adhE DNase I footprinting assay results showed that they shared a binding site (GTTCATTAATGAAC) in the adhE promoter. Overexpression and knockout of AcrR showed that AcrR was a novel regulator to promote the transcription of adhE The activator AcrR and repressor Rex may cross-regulate adhE when L. plantarum NF92 utilizes sorbitol or mannitol. Thus, a model of the control of adhE by AcrR and Rex during L. plantarum NF92 utilization of mannitol or sorbitol was proposed.IMPORTANCE The function and regulation of AdhE in the important probiotic genus Lactobacillus are rarely reported. Here we demonstrated that AdhE is responsible for sorbitol and mannitol utilization and is cross-regulated by two transcriptional regulators in L. plantarum NF92, which had not been reported previously. This is important for L. plantarum to compete and survive in some harsh environments in which sorbitol or mannitol could be used as carbon source. A novel transcriptional regulator AcrR was identified to be important to promote the expression of adhE, which was unknown before. The cross-regulation of adhE by AcrR and Rex is important to balance the level of NADH in the cell during sorbitol or mannitol utilization.
Keywords: DNA-binding regulator; Lactobacillus plantarum; aldehyde-alcohol dehydrogenase (AdhE); mannitol; sorbitol.
Copyright © 2019 American Society for Microbiology.
Figures


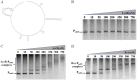



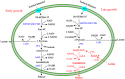

Similar articles
-
Transcriptional Regulator AcrR Increases Ethanol Tolerance through Regulation of Fatty Acid Synthesis in Lactobacillus plantarum.Appl Environ Microbiol. 2019 Oct 30;85(22):e01690-19. doi: 10.1128/AEM.01690-19. Print 2019 Nov 15. Appl Environ Microbiol. 2019. PMID: 31519657 Free PMC article.
-
High-level production of the low-calorie sugar sorbitol by Lactobacillus plantarum through metabolic engineering.Appl Environ Microbiol. 2007 Mar;73(6):1864-72. doi: 10.1128/AEM.02304-06. Epub 2007 Jan 19. Appl Environ Microbiol. 2007. PMID: 17261519 Free PMC article.
-
The bifunctional alcohol and aldehyde dehydrogenase gene, adhE, is necessary for ethanol production in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum.J Bacteriol. 2015 Apr;197(8):1386-93. doi: 10.1128/JB.02450-14. Epub 2015 Feb 9. J Bacteriol. 2015. PMID: 25666131 Free PMC article.
-
Ethanol catabolism in Aspergillus nidulans: a model system for studying gene regulation.Prog Nucleic Acid Res Mol Biol. 2001;69:149-204. doi: 10.1016/s0079-6603(01)69047-0. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11550794 Review.
-
Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer.Microb Cell Fact. 2011 Aug 30;10 Suppl 1(Suppl 1):S3. doi: 10.1186/1475-2859-10-S1-S3. Epub 2011 Aug 30. Microb Cell Fact. 2011. PMID: 21995294 Free PMC article. Review.
Cited by
-
Transcriptional Regulator AcrR Increases Ethanol Tolerance through Regulation of Fatty Acid Synthesis in Lactobacillus plantarum.Appl Environ Microbiol. 2019 Oct 30;85(22):e01690-19. doi: 10.1128/AEM.01690-19. Print 2019 Nov 15. Appl Environ Microbiol. 2019. PMID: 31519657 Free PMC article.
-
Differential Analysis of Stress Tolerance and Transcriptome of Probiotic Lacticaseibacillus casei Zhang Produced from Solid-State (SSF-SW) and Liquid-State (LSF-MRS) Fermentations.Microorganisms. 2020 Oct 26;8(11):1656. doi: 10.3390/microorganisms8111656. Microorganisms. 2020. PMID: 33114487 Free PMC article.
-
NAD+ pool depletion as a signal for the Rex regulon involved in Streptococcus agalactiae virulence.PLoS Pathog. 2021 Aug 9;17(8):e1009791. doi: 10.1371/journal.ppat.1009791. eCollection 2021 Aug. PLoS Pathog. 2021. PMID: 34370789 Free PMC article.
-
Illuminating the Genomic Landscape of Lactiplantibacillus plantarum PU3-A Novel Probiotic Strain Isolated from Human Breast Milk, Explored through Nanopore Sequencing.Microorganisms. 2023 Sep 28;11(10):2440. doi: 10.3390/microorganisms11102440. Microorganisms. 2023. PMID: 37894099 Free PMC article.
-
Role of the global regulator Rex in control of NAD+ -regeneration in Clostridioides (Clostridium) difficile.Mol Microbiol. 2019 Jun;111(6):1671-1688. doi: 10.1111/mmi.14245. Epub 2019 Apr 2. Mol Microbiol. 2019. PMID: 30882947 Free PMC article.
References
-
- Siezen RJ, Tzeneva VA, Castioni A, Wels M, Phan HT, Rademaker JL, Starrenburg MJ, Kleerebezem M, Molenaar D, van Hylckama Vlieg JE. 2010. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ Microbiol 12:758–773. doi:10.1111/j.1462-2920.2009.02119.x. - DOI - PubMed
-
- Martino ME, Bayjanov JR, Caffrey BE, Wels M, Joncour P, Hughes S, Gillet B, Kleerebezem M, van Hijum SA, Leulier F. 2016. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ Microbiol 18:4974–4989. doi:10.1111/1462-2920.13455. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

