Histone lysine dimethyl-demethylase KDM3A controls pathological cardiac hypertrophy and fibrosis
- PMID: 30531796
- PMCID: PMC6286331
- DOI: 10.1038/s41467-018-07173-2
Histone lysine dimethyl-demethylase KDM3A controls pathological cardiac hypertrophy and fibrosis
Abstract
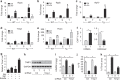
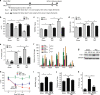
Left ventricular hypertrophy (LVH) is a major risk factor for cardiovascular morbidity and mortality. Pathological LVH engages transcriptional programs including reactivation of canonical fetal genes and those inducing fibrosis. Histone lysine demethylases (KDMs) are emerging regulators of transcriptional reprogramming in cancer, though their potential role in abnormal heart growth and fibrosis remains little understood. Here, we investigate gain and loss of function of an H3K9me2 specific demethylase, Kdm3a, and show it promotes LVH and fibrosis in response to pressure-overload. Cardiomyocyte KDM3A activates Timp1 transcription with pro-fibrotic activity. By contrast, a pan-KDM inhibitor, JIB-04, suppresses pressure overload-induced LVH and fibrosis. JIB-04 inhibits KDM3A and suppresses the transcription of fibrotic genes that overlap with genes downregulated in Kdm3a-KO mice versus WT controls. Our study provides genetic and biochemical evidence for a pro-hypertrophic function of KDM3A and proof-of principle for pharmacological targeting of KDMs as an effective strategy to counter LVH and pathological fibrosis.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA193455/CA/NCI NIH HHS/United States
- F31 HL134196/HL/NHLBI NIH HHS/United States
- T32 HL007227/HL/NHLBI NIH HHS/United States
- R01 HL128215/HL/NHLBI NIH HHS/United States
- R35 HL135827/HL/NHLBI NIH HHS/United States
- R01 CA125269/CA/NCI NIH HHS/United States
- R01 CA215063/CA/NCI NIH HHS/United States
- R01 HL120732/HL/NHLBI NIH HHS/United States
- R01 HL126012/HL/NHLBI NIH HHS/United States
- R01 HL119012/HL/NHLBI NIH HHS/United States
- R01 HL109471/HL/NHLBI NIH HHS/United States
- T35 HL134639/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

