Fungal secondary metabolism: regulation, function and drug discovery
- PMID: 30531948
- PMCID: PMC6381595
- DOI: 10.1038/s41579-018-0121-1
Fungal secondary metabolism: regulation, function and drug discovery
Abstract
One of the exciting movements in microbial sciences has been a refocusing and revitalization of efforts to mine the fungal secondary metabolome. The magnitude of biosynthetic gene clusters (BGCs) in a single filamentous fungal genome combined with the historic number of sequenced genomes suggests that the secondary metabolite wealth of filamentous fungi is largely untapped. Mining algorithms and scalable expression platforms have greatly expanded access to the chemical repertoire of fungal-derived secondary metabolites. In this Review, I discuss new insights into the transcriptional and epigenetic regulation of BGCs and the ecological roles of fungal secondary metabolites in warfare, defence and development. I also explore avenues for the identification of new fungal metabolites and the challenges in harvesting fungal-derived secondary metabolites.
Conflict of interest statement
Competing interests
There is no competing interest.
Figures

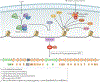
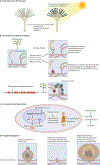


References
-
- Nesbitt BF, O’Kelly J, Sargeant K & Sheridan A Aspergillus flavus and turkey X disease: toxic metabolites of Aspergillus flavus. Nature 195, 1062–1063 (1962). - PubMed
-
-
Krause DJ et al. Functional and evolutionary characterization of a secondary metabolite gene cluster in budding yeasts. Proc. Natl Acad. Sci. USA 115, 11030–11035 (2018).
This study characterizes the pulcherrimin cluster in K. lactis, a yeast that belongs to a taxon not associated with secondary metabolism.
-
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

