Replication of MERS and SARS coronaviruses in bat cells offers insights to their ancestral origins
- PMID: 30531999
- PMCID: PMC6286955
- DOI: 10.1038/s41426-018-0208-9
Replication of MERS and SARS coronaviruses in bat cells offers insights to their ancestral origins
Abstract
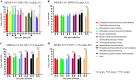
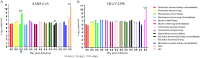
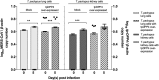
Previous findings of Middle East Respiratory Syndrome coronavirus (MERS-CoV)-related viruses in bats, and the ability of Tylonycteris-BatCoV HKU4 spike protein to utilize MERS-CoV receptor, human dipeptidyl peptidase 4 hDPP4, suggest a bat ancestral origin of MERS-CoV. We developed 12 primary bat cell lines from seven bat species, including Tylonycteris pachypus, Pipistrellus abramus and Rhinolophus sinicus (hosts of Tylonycteris-BatCoV HKU4, Pipistrellus-BatCoV HKU5, and SARS-related-CoV respectively), and tested their susceptibilities to MERS-CoVs, SARS-CoV, and human coronavirus 229E (HCoV-229E). Five cell lines, including P. abramus and R. sinicus but not T. pachypus cells, were susceptible to human MERS-CoV EMC/2012. However, three tested camel MERS-CoV strains showed different infectivities, with only two strains capable of infecting three and one cell lines respectively. SARS-CoV can only replicate in R. sinicus cells, while HCoV-229E cannot replicate in any bat cells. Bat dipeptidyl peptidase 4 (DPP4) sequences were closely related to those of human and non-human primates but distinct from dromedary DPP4 sequence. Critical residues for binding to MERS-CoV spike protein were mostly conserved in bat DPP4. DPP4 was expressed in the five bat cells susceptible to MERS-CoV, with significantly higher mRNA expression levels than those in non-susceptible cells (P = 0.0174), supporting that DPP4 expression is critical for MERS-CoV infection in bats. However, overexpression of T. pachypus DPP4 failed to confer MERS-CoV susceptibility in T. pachypus cells, suggesting other cellular factors in determining viral replication. The broad cellular tropism of MERS-CoV should prompt further exploration of host diversity of related viruses to identify its ancestral origin.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Rapid detection of MERS coronavirus-like viruses in bats: pote1ntial for tracking MERS coronavirus transmission and animal origin.Emerg Microbes Infect. 2018 Mar 7;7(1):18. doi: 10.1038/s41426-017-0016-7. Emerg Microbes Infect. 2018. PMID: 29511173 Free PMC article.
-
Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus.J Virol. 2013 Aug;87(15):8638-50. doi: 10.1128/JVI.01055-13. Epub 2013 May 29. J Virol. 2013. PMID: 23720729 Free PMC article.
-
Discovery of Novel Bat Coronaviruses in South China That Use the Same Receptor as Middle East Respiratory Syndrome Coronavirus.J Virol. 2018 Jun 13;92(13):e00116-18. doi: 10.1128/JVI.00116-18. Print 2018 Jul 1. J Virol. 2018. PMID: 29669833 Free PMC article.
-
Interspecies Jumping of Bat Coronaviruses.Viruses. 2021 Oct 29;13(11):2188. doi: 10.3390/v13112188. Viruses. 2021. PMID: 34834994 Free PMC article. Review.
-
Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond.Trends Microbiol. 2015 Aug;23(8):468-78. doi: 10.1016/j.tim.2015.06.003. Epub 2015 Jul 21. Trends Microbiol. 2015. PMID: 26206723 Free PMC article. Review.
Cited by
-
Entry of Scotophilus Bat Coronavirus-512 and Severe Acute Respiratory Syndrome Coronavirus in Human and Multiple Animal Cells.Pathogens. 2019 Nov 22;8(4):259. doi: 10.3390/pathogens8040259. Pathogens. 2019. PMID: 31766704 Free PMC article.
-
ACE2 from Pipistrellus abramus bats is a receptor for HKU5 coronaviruses.Nat Commun. 2025 May 28;16(1):4932. doi: 10.1038/s41467-025-60286-3. Nat Commun. 2025. PMID: 40436893 Free PMC article.
-
Management of epigenomic networks entailed in coronavirus infections and COVID-19.Clin Epigenetics. 2020 Aug 5;12(1):118. doi: 10.1186/s13148-020-00912-7. Clin Epigenetics. 2020. PMID: 32758273 Free PMC article. Review.
-
Animal models for the risk assessment of viral pandemic potential.Lab Anim Res. 2020 Apr 22;36:11. doi: 10.1186/s42826-020-00040-6. eCollection 2020. Lab Anim Res. 2020. PMID: 32337177 Free PMC article. Review.
-
[What do we know about the origin of COVID-19 three years later?].Rev Clin Esp. 2023 Apr;223(4):240-243. doi: 10.1016/j.rce.2023.02.002. Epub 2023 Mar 9. Rev Clin Esp. 2023. PMID: 37016626 Free PMC article. Review. Spanish.
References
-
- de Groot R. J. et al. Coronaviridae. In: Virus Taxonomy, Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses, International Union of Microbiological Societies, Virology Division, 806–828 (2011).
-
- Woo PC, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
