Cancer stem-like properties and gefitinib resistance are dependent on purine synthetic metabolism mediated by the mitochondrial enzyme MTHFD2
- PMID: 30532069
- PMCID: PMC6484769
- DOI: 10.1038/s41388-018-0589-1
Cancer stem-like properties and gefitinib resistance are dependent on purine synthetic metabolism mediated by the mitochondrial enzyme MTHFD2
Abstract
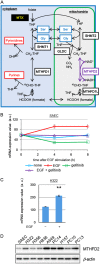
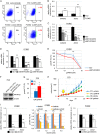
Tumor recurrence is attributable to cancer stem-like cells (CSCs), the metabolic mechanisms of which currently remain obscure. Here, we uncovered the critical role of folate-mediated one-carbon (1C) metabolism involving mitochondrial methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) and its downstream purine synthesis pathway. MTHFD2 knockdown greatly reduced tumorigenesis and stem-like properties, which were associated with purine nucleotide deficiency, and caused marked accumulation of 5-aminoimidazole carboxamide ribonucleotide (AICAR)-the final intermediate of the purine synthesis pathway. Lung cancer cells with acquired resistance to the targeted drug gefitinib, caused by elevated expression of components of the β-catenin pathway, exhibited increased stem-like properties and enhanced expression of MTHFD2. MTHFD2 knockdown or treatment with AICAR reduced the stem-like properties and restored gefitinib sensitivity in these gefitinib-resistant cancer cells. Moreover, overexpression of MTHFD2 in gefitinib-sensitive lung cancer cells conferred resistance to gefitinib. Thus, MTHFD2-mediated mitochondrial 1C metabolism appears critical for cancer stem-like properties and resistance to drugs including gefitinib through consumption of AICAR, leading to depletion of the intracellular pool of AICAR. Because CSCs are dependent on MTHFD2, therapies targeting MTHFD2 may eradicate tumors and prevent recurrence.
Conflict of interest statement
AT and NG obtained research funding from Daiichi Sankyo Co. ltd.
Figures




References
-
- Stewart BW, Wild C, International Agency for Research on Cancer, World Health Organization. World cancer report 2014. Lyon, France Geneva, Switzerland: International Agency for Research on Cancer WHO Press; 2014. xiv, p. 630.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

