Palmitoylation mediates membrane association of hepatitis E virus ORF3 protein and is required for infectious particle secretion
- PMID: 30532200
- PMCID: PMC6307819
- DOI: 10.1371/journal.ppat.1007471
Palmitoylation mediates membrane association of hepatitis E virus ORF3 protein and is required for infectious particle secretion
Abstract
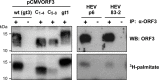
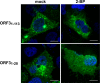
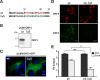
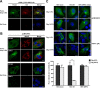
Hepatitis E virus (HEV) is a positive-strand RNA virus encoding 3 open reading frames (ORF). HEV ORF3 protein is a small, hitherto poorly characterized protein involved in viral particle secretion and possibly other functions. Here, we show that HEV ORF3 protein forms membrane-associated oligomers. Immunoblot analyses of ORF3 protein expressed in cell-free vs. cellular systems suggested a posttranslational modification. Further analyses revealed that HEV ORF3 protein is palmitoylated at cysteine residues in its N-terminal region, as corroborated by 3H-palmitate labeling, the investigation of cysteine-to-alanine substitution mutants and treatment with the palmitoylation inhibitor 2-bromopalmitate (2-BP). Abrogation of palmitoylation by site-directed mutagenesis or 2-BP treatment altered the subcellular localization of ORF3 protein, reduced the stability of the protein and strongly impaired the secretion of infectious particles. Moreover, selective membrane permeabilization coupled with immunofluorescence microscopy revealed that HEV ORF3 protein is entirely exposed to the cytosolic side of the membrane, allowing to propose a model for its membrane topology and interactions required in the viral life cycle. In conclusion, palmitoylation determines the subcellular localization, membrane topology and function of HEV ORF3 protein in the HEV life cycle.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

