Sequencing Analysis of Genetic Loci for Resistance for Late Leaf Spot and Rust in Peanut (Arachis hypogaea L.)
- PMID: 30534132
- PMCID: PMC6275244
- DOI: 10.3389/fpls.2018.01727
Sequencing Analysis of Genetic Loci for Resistance for Late Leaf Spot and Rust in Peanut (Arachis hypogaea L.)
Abstract
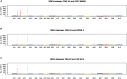
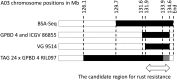
The aim of this study was to identify candidate resistance genes for late leaf spot (LLS) and rust diseases in peanut (Arachis hypogaea L.). We used a double-digest restriction-site associated DNA sequencing (ddRAD-Seq) technique based on next-generation sequencing (NGS) for genotyping analysis across the recombinant inbred lines (RILs) derived from a cross between a susceptible line, TAG 24, and a resistant line, GPBD 4. A total of 171 SNPs from the ddRAD-Seq together with 282 markers published in the previous studies were mapped on a genetic map covering 1510.1 cM. Subsequent quantitative trait locus (QTL) analysis revealed major genetic loci for LLS and rust resistance on chromosomes A02 and A03, respectively. Heterogeneous inbred family-derived near isogenic lines and the pedigree of the resistant gene donor, A. cardenasii Krapov. & W.C. Greg., including the resistant derivatives of ICGV 86855 and VG 9514 as well as GPBD 4, were employed for whole-genome resequencing analysis. The results indicated the QTL candidates for LLS and rust resistance were located in 1.4- and 2.7-Mb genome regions on A02 and A03, respectively. In these regions, four and six resistance-related genes with deleterious mutations were selected as candidates for LLS and rust resistance, respectively. These delimited genomic regions may be beneficial in breeding programs aimed at improving disease resistance and enhancing peanut productivity.
Keywords: late leaf spot and rust diseases; peanut; quantitative trait locus; restriction-site associated DNA sequencing; whole-genome resequencing analysis.
Figures

References
-
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., Wang L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6 80–92. 10.4161/fly.19695 - DOI - PMC - PubMed
-
- Clevenger J., Bertioli D. J., Leal-Bertioli S. C. M., Chu Y., Stalker H. T., Ozias-Akins P. (2017). IntroMap: a pipeline and set of diagnostic diploid Arachis SNPs as a tool for mapping alien introgressions in Arachis hypogaea. Peanut Sci. 44 66–73. 10.3146/PS17-5.1 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

