Dissecting the Genomic Architecture of Resistance to Eimeria maxima Parasitism in the Chicken
- PMID: 30534137
- PMCID: PMC6275401
- DOI: 10.3389/fgene.2018.00528
Dissecting the Genomic Architecture of Resistance to Eimeria maxima Parasitism in the Chicken
Abstract
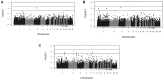
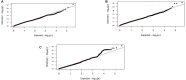
Coccidiosis in poultry, caused by protozoan parasites of the genus Eimeria, is an intestinal disease with substantial economic impact. With the use of anticoccidial drugs under public and political pressure, and the comparatively higher cost of live-attenuated vaccines, an attractive complementary strategy for control is to breed chickens with increased resistance to Eimeria parasitism. Prior infection with Eimeria maxima leads to complete immunity against challenge with homologous strains, but only partial resistance to challenge with antigenically diverse heterologous strains. We investigate the genetic architecture of avian resistance to E. maxima primary infection and heterologous strain secondary challenge using White Leghorn populations of derived inbred lines, C.B12 and 15I, known to differ in susceptibility to the parasite. An intercross population was infected with E. maxima Houghton (H) strain, followed 3 weeks later by E. maxima Weybridge (W) strain challenge, while a backcross population received a single E. maxima W infection. The phenotypes measured were parasite replication (counting fecal oocyst output or qPCR for parasite numbers in intestinal tissue), intestinal lesion score (gross pathology, scale 0-4), and for the backcross only, serum interleukin-10 (IL-10) levels. Birds were genotyped using a high density genome-wide DNA array (600K, Affymetrix). Genome-wide association study located associations on chromosomes 1, 2, 3, and 5 following primary infection in the backcross population, and a suggestive association on chromosome 1 following heterologous E. maxima W challenge in the intercross population. This mapped several megabases away from the quantitative trait locus (QTL) linked to the backcross primary W strain infection, suggesting different underlying mechanisms for the primary- and heterologous secondary- responses. Underlying pathways for those genes located in the respective QTL for resistance to primary infection and protection against heterologous challenge were related mainly to immune response, with IL-10 signaling in the backcross primary infection being the most significant. Additionally, the identified markers associated with IL-10 levels exhibited significant additive genetic variance. We suggest this is a phenotype of interest to the outcome of challenge, being scalable in live birds and negating the requirement for single-bird cages, fecal oocyst counts, or slaughter for sampling (qPCR).
Keywords: Eimeria maxima; QTL; backcross; intercross; interleukin-10; oocyst output; resistance.
Figures






References
-
- Blake D. P., Hesketh P., Archer A., Carroll F., Smith A. L., Shirley M. W. (2004). Parasite genetics and the immune host: recombination between antigenic types of Eimeria maxima as an entrée to the identification of protective antigens. Mol. Biochem. Parasitol. 138 143–152. 10.1016/j.molbiopara.2004.08.006 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

