Development of multiplex PCR and multi-color fluorescent in situ hybridization (m-FISH) coupled protocol for detection and imaging of multi-pathogens involved in inflammatory bowel disease
- PMID: 30534203
- PMCID: PMC6280354
- DOI: 10.1186/s13099-018-0278-1
Development of multiplex PCR and multi-color fluorescent in situ hybridization (m-FISH) coupled protocol for detection and imaging of multi-pathogens involved in inflammatory bowel disease
Abstract
Background: Several pathogens have been debated to play a role in inflammatory bowel disease (IBD) including Crohn's disease (CD). None of these pathogens have been investigated together in same clinical samples. We developed a multiplex PCR and multi-color fluorescent in situ hybridization (m-FISH) protocols for simultaneous detection of CD-associated pathogens including Mycobacterium avium subspecies paratuberculosis (MAP), Klebsiella pneumoniae, and adherent-invasive Escherichia coli strain LF82.
Methods: The multiplex PCR is based on 1-h DNAzol® extraction protocol modified for rapid extraction of bacterial DNA from culture, blood, and intestinal biopsies. Oligonucleotide primers sequences unique to these pathogens were evaluated individually and in combinations using bioinformatics and experimental approaches. m-FISH was based on fluorescent-tagged oligonucleotides and confocal scanning laser microscopy (CSLM).
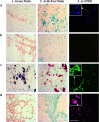
Results: Following several attempts, the concentration of the oligonucleotide primers and DNA templates and the PCR annealing temperatures were optimized. Multiplex PCR analyses revealed excellent amplification signal in trials where a single primer set and combinations of two and three primers sets were tested against a mixture of DNA from three different bacteria or a mixture of three bacterial cultures mixed in one tube before DNA extraction. Slides with individual and mixtures of bacterial cultures and intestinal tissue sections from IBD patients were tested by m-FISH and the CSLM images verified multiplex PCR results detected on 3% agarose gel.
Conclusion: We developed a 4-h multiplex PCR protocol, which was validated by m-FISH images, capable of detecting up to four genes from major pathogens associated with CD. The new protocol should serve as an excellent tool to support efforts to study multi-pathogens involved in CD and other autoimmune disease.
Keywords: Crohn’s disease; MAP; Multiplex PCR; Ulcerative colitis; m-FISH.
Figures





References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

