The draft genomes of five agriculturally important African orphan crops
- PMID: 30535374
- PMCID: PMC6405277
- DOI: 10.1093/gigascience/giy152
The draft genomes of five agriculturally important African orphan crops
Abstract
Background: The expanding world population is expected to double the worldwide demand for food by 2050. Eighty-eight percent of countries currently face a serious burden of malnutrition, especially in Africa and south and southeast Asia. About 95% of the food energy needs of humans are fulfilled by just 30 species, of which wheat, maize, and rice provide the majority of calories. Therefore, to diversify and stabilize the global food supply, enhance agricultural productivity, and tackle malnutrition, greater use of neglected or underutilized local plants (so-called orphan crops, but also including a few plants of special significance to agriculture, agroforestry, and nutrition) could be a partial solution.
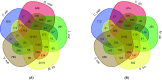
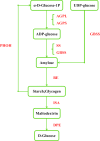
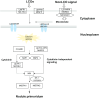
Results: Here, we present draft genome information for five agriculturally, biologically, medicinally, and economically important underutilized plants native to Africa: Vigna subterranea, Lablab purpureus, Faidherbia albida, Sclerocarya birrea, and Moringa oleifera. Assembled genomes range in size from 217 to 654 Mb. In V. subterranea, L. purpureus, F. albida, S. birrea, and M. oleifera, we have predicted 31,707, 20,946, 28,979, 18,937, and 18,451 protein-coding genes, respectively. By further analyzing the expansion and contraction of selected gene families, we have characterized root nodule symbiosis genes, transcription factors, and starch biosynthesis-related genes in these genomes.
Conclusions: These genome data will be useful to identify and characterize agronomically important genes and understand their modes of action, enabling genomics-based, evolutionary studies, and breeding strategies to design faster, more focused, and predictable crop improvement programs.
Keywords: food security; orphan crops; root nodule symbiosis; transcription factor; transcriptome; whole-genome sequencing.
© The Author(s) 2018. Published by Oxford University Press.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Comment in
-
Sequencing forgotten crops.Nat Plants. 2019 Jan;5(1):5. doi: 10.1038/s41477-018-0354-z. Nat Plants. 2019. PMID: 30626921 No abstract available.
References
-
- United Nations, Department of Economic and Social Affairs, Population Division. World population prospects: the 2017 revision, Key Findings and Advance Tables. 2017. Working Paper No. ESA/P/WP/248.
-
- Development Initiatives. Global nutrition report 2017: nourishing the SDGs. Bristol, UK: Development Initiatives; 2017.
-
- Allender C. The second report on the state of the world's plant genetic resources for food and agriculture. Experimental Agriculture. 2011;47(3):574–574.
-
- African Orphan Crops Consortium: http://www.africanorphancrops.org (2018). Accessed 20 Nov 2018.
-
- Varshney RK, Chen W, Li Yet al., Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol. 2011;30:83–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

