Resistance to Epigenetic-Targeted Therapy Engenders Tumor Cell Vulnerabilities Associated with Enhancer Remodeling
- PMID: 30537514
- PMCID: PMC6352909
- DOI: 10.1016/j.ccell.2018.11.005
Resistance to Epigenetic-Targeted Therapy Engenders Tumor Cell Vulnerabilities Associated with Enhancer Remodeling
Abstract
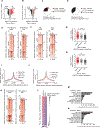
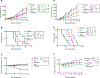
Drug resistance represents a major challenge to achieving durable responses to cancer therapeutics. Resistance mechanisms to epigenetically targeted drugs remain largely unexplored. We used bromodomain and extra-terminal domain (BET) inhibition in neuroblastoma as a prototype to model resistance to chromatin modulatory therapeutics. Genome-scale, pooled lentiviral open reading frame (ORF) and CRISPR knockout rescue screens nominated the phosphatidylinositol 3-kinase (PI3K) pathway as promoting resistance to BET inhibition. Transcriptomic and chromatin profiling of resistant cells revealed that global enhancer remodeling is associated with upregulation of receptor tyrosine kinases (RTKs), activation of PI3K signaling, and vulnerability to RTK/PI3K inhibition. Large-scale combinatorial screening with BET inhibitors identified PI3K inhibitors among the most synergistic upfront combinations. These studies provide a roadmap to elucidate resistance to epigenetic-targeted therapeutics and inform efficacious combination therapies.
Keywords: BET inhibition; MYCN; PI3K signaling; drug resistance; enhancer remodeling; neuroblastoma.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
K.S. participates in the DFCI/Novartis Drug Discovery Program which includes grant support for an unrelated project and previously included consulting and has consulted for Rigel Pharmaceuticals on a topic unrelated to this manuscript. R.B. and P.B. receive grant funding from Novartis Institute for Biomedical Research for unrelated projects. W.A.W. is founder of StemSynergy Therapeutics which works on targeting WNT signaling in colorectal cancer.
Figures





References
-
- Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, Taussig DC, Rezai K, Roumier C, Herait P, et al. (2016). Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. The Lancet Haematology 3, e186–195. - PubMed
-
- Boeva V, Louis-Brennetot C, Peltier A, Durand S, Pierre-Eugene C, Raynal V, Etchevers HC, Thomas S, Lermine A, Daudigeos-Dubus E, et al. (2017). Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nature genetics. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

