A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1
- PMID: 30537986
- PMCID: PMC6290540
- DOI: 10.1186/s13059-018-1594-y
A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1
Abstract
Background: Friend leukemia virus integration 1 (FLI1), an ETS transcription factor family member, acts as an oncogenic driver in hematological malignancies and promotes tumor growth in solid tumors. However, little is known about the mechanisms underlying the activation of this proto-oncogene in tumors.
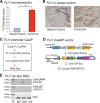
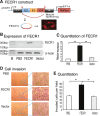
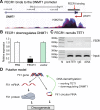
Results: Immunohistochemical staining showed that FLI1 is aberrantly overexpressed in advanced stage and metastatic breast cancers. Using a CRISPR Cas9-guided immunoprecipitation assay, we identify a circular RNA in the FLI1 promoter chromatin complex, consisting of FLI1 exons 4-2-3, referred to as FECR1.Overexpression of FECR1 enhances invasiveness of MDA-MB231 breast cancer cells. Notably, FECR1 utilizes a positive feedback mechanism to activate FLI1 by inducing DNA hypomethylation in CpG islands of the promoter. FECR1 binds to the FLI1 promoter in cis and recruits TET1, a demethylase that is actively involved in DNA demethylation. FECR1 also binds to and downregulates in trans DNMT1, a methyltransferase that is essential for the maintenance of DNA methylation.
Conclusions: These data suggest that FECR1 circular RNA acts as an upstream regulator to control breast cancer tumor growth by coordinating the regulation of DNA methylating and demethylating enzymes. Thus, FLI1 drives tumor metastasis not only through the canonical oncoprotein pathway, but also by using epigenetic mechanisms mediated by its exonic circular RNA.
Keywords: Breast cancer; Circular RNA; DNA methylation; DNMT1; FLI1; TET1; Tumor.
Conflict of interest statement
Ethics approval and consent to participate
The study was conducted in compliance with the Helsinki Declaration. The study protocol was approved by the Research Ethics Board of the First Hospital of Jilin University (IRB 2015-260). Informed consent was obtained from each participant in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

