Systematic deletions in the cellobiohydrolase (CBH) Cel7A from the fungus Trichoderma reesei reveal flexible loops critical for CBH activity
- PMID: 30538133
- PMCID: PMC6369288
- DOI: 10.1074/jbc.RA118.006699
Systematic deletions in the cellobiohydrolase (CBH) Cel7A from the fungus Trichoderma reesei reveal flexible loops critical for CBH activity
Abstract
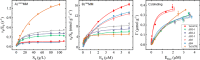
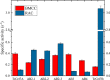
Glycoside hydrolase family 7 (GH7) cellulases are some of the most efficient degraders of cellulose, making them particularly relevant for industries seeking to produce renewable fuels from lignocellulosic biomass. The secretome of the cellulolytic model fungus Trichoderma reesei contains two GH7s, termed TrCel7A and TrCel7B. Despite having high structural and sequence similarities, the two enzymes are functionally quite different. TrCel7A is an exolytic, processive cellobiohydrolase (CBH), with high activity on crystalline cellulose, whereas TrCel7B is an endoglucanase (EG) with a preference for more amorphous cellulose. At the structural level, these functional differences are usually ascribed to the flexible loops that cover the substrate-binding areas. TrCel7A has an extensive tunnel created by eight peripheral loops, and the absence of four of these loops in TrCel7B makes its catalytic domain a more open cleft. To investigate the structure-function relationships of these loops, here we produced and kinetically characterized several variants in which four loops unique to TrCel7A were individually deleted to resemble the arrangement in the TrCel7B structure. Analysis of a range of kinetic parameters consistently indicated that the B2 loop, covering the substrate-binding subsites -3 and -4 in TrCel7A, was a key determinant for the difference in CBH- or EG-like behavior between TrCel7A and TrCel7B. Conversely, the B3 and B4 loops, located closer to the catalytic site in TrCel7A, were less important for these activities. We surmise that these results could be useful both in further mechanistic investigations and for guiding engineering efforts of this industrially important enzyme family.
Keywords: Cel7A; Cel7B; Trichoderma reesei; cellobiohydrolase; cellulase; cellulose; endoglucanase; enzyme kinetics; loop engineering; protein engineering.
© 2019 Schiano-di-Cola et al.
Conflict of interest statement
K. J. and K. B. are employed by Novozymes, a major enzyme-producing company
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

