Crystal structure of the human NK1 tachykinin receptor
- PMID: 30538204
- PMCID: PMC6310836
- DOI: 10.1073/pnas.1812717115
Crystal structure of the human NK1 tachykinin receptor
Abstract
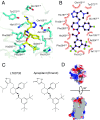
The NK1 tachykinin G-protein-coupled receptor (GPCR) binds substance P, the first neuropeptide to be discovered in mammals. Through activation of NK1R, substance P modulates a wide variety of physiological and disease processes including nociception, inflammation, and depression. Human NK1R (hNK1R) modulators have shown promise in clinical trials for migraine, depression, and emesis. However, the only currently approved drugs targeting hNK1R are inhibitors for chemotherapy-induced nausea and vomiting (CINV). To better understand the molecular basis of ligand recognition and selectivity, we solved the crystal structure of hNK1R bound to the inhibitor L760735, a close analog of the drug aprepitant. Our crystal structure reveals the basis for antagonist interaction in the deep and narrow orthosteric pocket of the receptor. We used our structure as a template for computational docking and molecular-dynamics simulations to dissect the energetic importance of binding pocket interactions and model the binding of aprepitant. The structure of hNK1R is a valuable tool in the further development of tachykinin receptor modulators for multiple clinical applications.
Keywords: GPCR; drug design; ligand recognition; substance P; tachykinin receptor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Michelot R, Mayer M, Magneney S, Thierry J, Potier P. Substance P and Neurokinins. Springer; New York: 1987. Activity of the C-terminal part in tachykinins on guinea pig ileum and trachea preparations; pp. 158–160.
-
- Almeida TA, et al. Tachykinins and tachykinin receptors: Structure and activity relationships. Curr Med Chem. 2004;11:2045–2081. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

