Social status alters chromatin accessibility and the gene regulatory response to glucocorticoid stimulation in rhesus macaques
- PMID: 30538209
- PMCID: PMC6347725
- DOI: 10.1073/pnas.1811758115
Social status alters chromatin accessibility and the gene regulatory response to glucocorticoid stimulation in rhesus macaques
Abstract
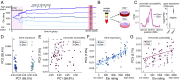
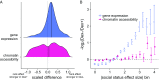
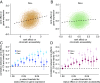
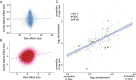
Low social status is an important predictor of disease susceptibility and mortality risk in humans and other social mammals. These effects are thought to stem in part from dysregulation of the glucocorticoid (GC)-mediated stress response. However, the molecular mechanisms that connect low social status and GC dysregulation to downstream health outcomes remain elusive. Here, we used an in vitro GC challenge to investigate the consequences of experimentally manipulated social status (i.e., dominance rank) for immune cell gene regulation in female rhesus macaques, using paired control and GC-treated peripheral blood mononuclear cell samples. We show that social status not only influences immune cell gene expression but also chromatin accessibility at hundreds of regions in the genome. Social status effects on gene expression were less pronounced following GC treatment than under control conditions. In contrast, social status effects on chromatin accessibility were stable across conditions, resulting in an attenuated relationship between social status, chromatin accessibility, and gene expression after GC exposure. Regions that were more accessible in high-status animals and regions that become more accessible following GC treatment were enriched for a highly concordant set of transcription factor binding motifs, including motifs for the GC receptor cofactor AP-1. Together, our findings support the hypothesis that social status alters the dynamics of GC-mediated gene regulation and identify chromatin accessibility as a mechanism involved in social stress-driven GC resistance. More broadly, they emphasize the context-dependent nature of social status effects on gene regulation and implicate epigenetic remodeling of chromatin accessibility as a contributing factor.
Keywords: chromatin accessibility; dominance rank; epigenomics; gene regulation; social status.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
Molecular origins and outcomes of status and stress in primates.Proc Natl Acad Sci U S A. 2019 Jan 22;116(4):1081-1083. doi: 10.1073/pnas.1819472116. Epub 2019 Jan 14. Proc Natl Acad Sci U S A. 2019. PMID: 30642957 Free PMC article. No abstract available.
References
-
- Marmot MG, Sapolsky R. Of baboons and men: Social circumstances, biology, and the social gradient in health. In: Weinstein M, Lane MA, editors. Sociality, Hierarchy, Health: Comparative Biodemography. National Academies Press; Washington, DC: 2014. pp. 365–388. - PubMed
-
- Marmot MG. The Status Syndrome: How Social Standing Affects our Health and Longevity. Macmillan; New York: 2004.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

