Histone demethylase LSD1 is required for germinal center formation and BCL6-driven lymphomagenesis
- PMID: 30538335
- PMCID: PMC6294324
- DOI: 10.1038/s41590-018-0273-1
Histone demethylase LSD1 is required for germinal center formation and BCL6-driven lymphomagenesis
Abstract
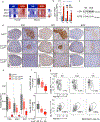
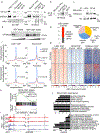
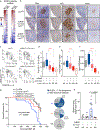
Germinal center (GC) B cells feature repression of many gene enhancers to establish their characteristic transcriptome. Here we show that conditional deletion of Lsd1 in GCs significantly impaired GC formation, associated with failure to repress immune synapse genes linked to GC exit, which are also direct targets of the transcriptional repressor BCL6. We found that BCL6 directly binds LSD1 and recruits it primarily to intergenic and intronic enhancers. Conditional deletion of Lsd1 suppressed GC hyperplasia caused by constitutive expression of BCL6 and significantly delayed BCL6-driven lymphomagenesis. Administration of catalytic inhibitors of LSD1 had little effect on GC formation or GC-derived lymphoma cells. Using a CRISPR-Cas9 domain screen, we found instead that the LSD1 Tower domain was critical for dependence on LSD1 in GC-derived B cells. These results indicate an essential role for LSD1 in the humoral immune response, where it modulates enhancer function by forming repression complexes with BCL6.
Figures



Comment in
-
B cells turn on, tune in with LSD1.Nat Immunol. 2019 Jan;20(1):3-5. doi: 10.1038/s41590-018-0281-1. Nat Immunol. 2019. PMID: 30538329 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

