Ancestrality and Mosaicism of Giant Viruses Supporting the Definition of the Fourth TRUC of Microbes
- PMID: 30538677
- PMCID: PMC6277510
- DOI: 10.3389/fmicb.2018.02668
Ancestrality and Mosaicism of Giant Viruses Supporting the Definition of the Fourth TRUC of Microbes
Abstract
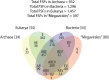
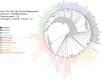
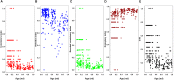
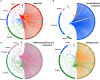
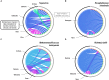
Giant viruses of amoebae were discovered in 2003. Since then, their diversity has greatly expanded. They were suggested to form a fourth branch of life, collectively named 'TRUC' (for "Things Resisting Uncompleted Classifications") alongside Bacteria, Archaea, and Eukarya. Their origin and ancestrality remain controversial. Here, we specify the evolution and definition of giant viruses. Phylogenetic and phenetic analyses of informational gene repertoires of giant viruses and selected bacteria, archaea and eukaryota were performed, including structural phylogenomics based on protein structural domains grouped into 289 universal fold superfamilies (FSFs). Hierarchical clustering analysis was performed based on a binary presence/absence matrix constructed using 727 informational COGs from cellular organisms. The presence/absence of 'universal' FSF domains was used to generate an unrooted maximum parsimony phylogenomic tree. Comparison of the gene content of a giant virus with those of a bacterium, an archaeon, and a eukaryote with small genomes was also performed. Overall, both cladistic analyses based on gene sequences of very central and ancient proteins and on highly conserved protein fold structures as well as phenetic analyses were congruent regarding the delineation of a fourth branch of microbes comprised by giant viruses. Giant viruses appeared as a basal group in the tree of all proteomes. A pangenome and core genome determined for Rickettsia bellii (bacteria), Methanomassiliicoccus luminyensis (archaeon), Encephalitozoon intestinalis (eukaryote), and Tupanvirus (giant virus) showed a substantial proportion of Tupanvirus genes that overlap with those of the cellular microbes. In addition, a substantial genome mosaicism was observed, with 51, 11, 8, and 0.2% of Tupanvirus genes best matching with viruses, eukaryota, bacteria, and archaea, respectively. Finally, we found that genes themselves may be subject to lateral sequence transfers. In summary, our data highlight the quantum leap between classical and giant viruses. Phylogenetic and phyletic analyses and the study of protein fold superfamilies confirm previous evidence of the existence of a fourth TRUC of life that includes giant viruses, and highlight its ancestrality and mosaicism. They also point out that best evolutionary representations for giant viruses and cellular microorganisms are rhizomes, and that sequence transfers rather than gene transfers have to be considered.
Keywords: TRUC; giant virus; informational genes; megavirales; mimivirus; protein structural domains.
Figures






References
-
- Abrahao J., Silva L., Santos Silva L., Bou Khalil J. Y., Rodrigues R., Arantes T., et al. (2018). Tupanvirus, a tailed giant virus and distant relative of Mimiviridae, possesses the most complete translational apparatus of the virosphere. Nat. Commun. 9:749. 10.1038/s41467-018-03168-1 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

