A New Phylogenetic Framework for the Animal-Adapted Mycobacterium tuberculosis Complex
- PMID: 30538680
- PMCID: PMC6277475
- DOI: 10.3389/fmicb.2018.02820
A New Phylogenetic Framework for the Animal-Adapted Mycobacterium tuberculosis Complex
Abstract
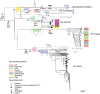
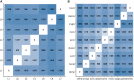
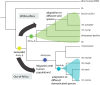
Tuberculosis (TB) affects humans and other animals and is caused by bacteria from the Mycobacterium tuberculosis complex (MTBC). Previous studies have shown that there are at least nine members of the MTBC infecting animals other than humans; these have also been referred to as ecotypes. However, the ecology and the evolution of these animal-adapted MTBC ecotypes are poorly understood. Here we screened 12,886 publicly available MTBC genomes and newly sequenced 17 animal-adapted MTBC strains, gathering a total of 529 genomes of animal-adapted MTBC strains. Phylogenomic and comparative analyses confirm that the animal-adapted MTBC members are paraphyletic with some members more closely related to the human-adapted Mycobacterium africanum Lineage 6 than to other animal-adapted strains. Furthermore, we identified four main animal-adapted MTBC clades that might correspond to four main host shifts; two of these clades are hypothesized to reflect independent cattle domestication events. Contrary to what would be expected from an obligate pathogen, MTBC nucleotide diversity was not positively correlated with host phylogenetic distances, suggesting that host tropism in the animal-adapted MTBC seems to be driven by contact rates and demographic aspects of the host population rather by than host relatedness. By combining phylogenomics with ecological data, we propose an evolutionary scenario in which the ancestor of Lineage 6 and all animal-adapted MTBC ecotypes was a generalist pathogen that subsequently adapted to different host species. This study provides a new phylogenetic framework to better understand the evolution of the different ecotypes of the MTBC and guide future work aimed at elucidating the molecular mechanisms underlying host range.
Keywords: genetic diversity; host range; host–pathogen interactions; specificity; whole-genome sequencing.
Figures



References
-
- Ates L. S., Sayes F., Frigui W., Ummels R., Damen M. P. M., Bottai D., et al. (2018b). RD5-mediated lack of PE_PGRS and PPE-MPTR export in BCG vaccine strains results in strong reduction of antigenic repertoire but little impact on protection. PLoS Pathog. 14:e1007139. 10.1371/journal.ppat.1007139 - DOI - PMC - PubMed

