A robust pipeline with high replication rate for detection of somatic variants in the adaptive immune system as a source of common genetic variation in autoimmune disease
- PMID: 30541027
- PMCID: PMC6452186
- DOI: 10.1093/hmg/ddy425
A robust pipeline with high replication rate for detection of somatic variants in the adaptive immune system as a source of common genetic variation in autoimmune disease
Abstract
The role of somatic variants in diseases beyond cancer is increasingly being recognized, with potential roles in autoinflammatory and autoimmune diseases. However, as mutation rates and allele fractions are lower, studies in these diseases are substantially less tolerant of false positives, and bio-informatics algorithms require high replication rates. We developed a pipeline combining two variant callers, MuTect2 and VarScan2, with technical filtering and prioritization. Our pipeline detects somatic variants with allele fractions as low as 0.5% and achieves a replication rate of >55%. Validation in an independent data set demonstrates excellent performance (sensitivity > 57%, specificity > 98%, replication rate > 80%). We applied this pipeline to the autoimmune disease multiple sclerosis (MS) as a proof-of-principle. We demonstrate that 60% of MS patients carry 2-10 exonic somatic variants in their peripheral blood T and B cells, with the vast majority (80%) occurring in T cells and variants persisting over time. Synonymous variants significantly co-occur with non-synonymous variants. Systematic characterization indicates somatic variants are enriched for being novel or very rare in public databases of germline variants and trend towards being more damaging and conserved, as reflected by higher phred-scaled combined annotation-dependent depletion (CADD) and genomic evolutionary rate profiling (GERP) scores. Our pipeline and proof-of-principle now warrant further investigation of common somatic genetic variation on top of inherited genetic variation in the context of autoimmune disease, where it may offer subtle survival advantages to immune cells and contribute to the capacity of these cells to participate in the autoimmune reaction.
© The Author(s) 2018. Published by Oxford University Press.
Figures

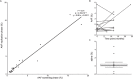

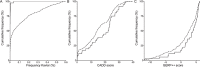
Similar articles
-
SNooPer: a machine learning-based method for somatic variant identification from low-pass next-generation sequencing.BMC Genomics. 2016 Nov 14;17(1):912. doi: 10.1186/s12864-016-3281-2. BMC Genomics. 2016. PMID: 27842494 Free PMC article.
-
Optimized pipeline of MuTect and GATK tools to improve the detection of somatic single nucleotide polymorphisms in whole-exome sequencing data.BMC Bioinformatics. 2016 Nov 8;17(Suppl 12):341. doi: 10.1186/s12859-016-1190-7. BMC Bioinformatics. 2016. PMID: 28185561 Free PMC article.
-
pyAmpli: an amplicon-based variant filter pipeline for targeted resequencing data.BMC Bioinformatics. 2017 Dec 14;18(1):554. doi: 10.1186/s12859-017-1985-1. BMC Bioinformatics. 2017. PMID: 29237398 Free PMC article.
-
Detecting pathogenic variants in autoimmune diseases using high-throughput sequencing.Immunol Cell Biol. 2021 Feb;99(2):146-156. doi: 10.1111/imcb.12372. Epub 2020 Jul 27. Immunol Cell Biol. 2021. PMID: 32623783 Free PMC article. Review.
-
Understanding the limitations of next generation sequencing informatics, an approach to clinical pipeline validation using artificial data sets.Cancer Genet. 2013 Dec;206(12):441-8. doi: 10.1016/j.cancergen.2013.11.005. Epub 2013 Nov 28. Cancer Genet. 2013. PMID: 24528889 Review.
Cited by
-
Multiple Sclerosis-Associated hnRNPA1 Mutations Alter hnRNPA1 Dynamics and Influence Stress Granule Formation.Int J Mol Sci. 2021 Mar 12;22(6):2909. doi: 10.3390/ijms22062909. Int J Mol Sci. 2021. PMID: 33809384 Free PMC article.
-
A targeted sequencing extension for transcript genotyping in single-cell transcriptomics.Life Sci Alliance. 2023 Sep 11;6(11):e202301971. doi: 10.26508/lsa.202301971. Print 2023 Nov. Life Sci Alliance. 2023. PMID: 37696578 Free PMC article.
-
Design of Personalized Neoantigen RNA Vaccines Against Cancer Based on Next-Generation Sequencing Data.Methods Mol Biol. 2022;2547:165-185. doi: 10.1007/978-1-0716-2573-6_7. Methods Mol Biol. 2022. PMID: 36068464
-
Adult-Onset Anti-Citrullinated Peptide Antibody-Negative Destructive Rheumatoid Arthritis Is Characterized by a Disease-Specific CD8+ T Lymphocyte Signature.Front Immunol. 2020 Nov 19;11:578848. doi: 10.3389/fimmu.2020.578848. eCollection 2020. Front Immunol. 2020. PMID: 33329548 Free PMC article.
-
Clonal hematopoiesis, somatic mosaicism, and age-associated disease.Physiol Rev. 2023 Jan 1;103(1):649-716. doi: 10.1152/physrev.00004.2022. Epub 2022 Sep 1. Physiol Rev. 2023. PMID: 36049115 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

