A nearest-neighbors network model for sequence data reveals new insight into genotype distribution of a pathogen
- PMID: 30541438
- PMCID: PMC6291930
- DOI: 10.1186/s12859-018-2453-2
A nearest-neighbors network model for sequence data reveals new insight into genotype distribution of a pathogen
Abstract
Background: Sequence similarity networks are useful for classifying and characterizing biologically important proteins. Threshold-based approaches to similarity network construction using exact distance measures are prohibitively slow to compute and rely on the difficult task of selecting an appropriate threshold, while similarity networks based on approximate distance calculations compromise useful structural information.
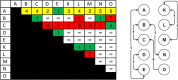
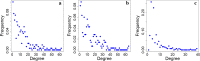
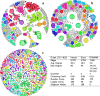
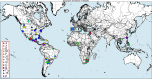
Results: We present an alternative network representation for a set of sequence data that overcomes these drawbacks. In our model, called the Directed Weighted All Nearest Neighbors (DiWANN) network, each sequence is represented by a node and is connected via a directed edge to only the closest sequence, or sequences in the case of ties, in the dataset. Our contributions span several aspects. Specifically, we: (i) Apply an all nearest neighbors network model to protein sequence data from three different applications and examine the structural properties of the networks; (ii) Compare the model against threshold-based networks to validate their semantic equivalence, and demonstrate the relative advantages the model offers; (iii) Demonstrate the model's resilience to missing sequences; and (iv) Develop an efficient algorithm for constructing a DiWANN network from a set of sequences. We find that the DiWANN network representation attains similar semantic properties to threshold-based graphs, while avoiding weaknesses of both high and low threshold graphs. Additionally, we find that approximate distance networks, using BLAST bitscores in place of exact edit distances, can cause significant loss of structural information. We show that the proposed DiWANN network construction algorithm provides a fourfold speedup over a standard threshold based approach to network construction. We also identify a relationship between the centrality of a sequence in a similarity network of an Anaplasma marginale short sequence repeat dataset and how broadly that sequence is dispersed geographically.
Conclusion: We demonstrate that using approximate distance measures to rapidly construct similarity networks may lead to significant deficiencies in the structure of that network in terms centrality and clustering analyses. We present a new network representation that maintains the structural semantics of threshold-based networks while increasing connectedness, and an algorithm for constructing the network using exact distance measures in a fraction of the time it would take to build a threshold-based equivalent.
Keywords: Anaplasma marginale Msp1a; Centrality; Clustering; GroEL; Network analysis; Sequence similarity network.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures







Similar articles
-
Sequence Similarity Network Analysis Provides Insight into the Temporal and Geographical Distribution of Mutations in SARS-CoV-2 Spike Protein.Viruses. 2022 Jul 29;14(8):1672. doi: 10.3390/v14081672. Viruses. 2022. PMID: 36016294 Free PMC article.
-
Network analysis of driver genes in human cancers.Front Bioinform. 2024 Jul 8;4:1365200. doi: 10.3389/fbinf.2024.1365200. eCollection 2024. Front Bioinform. 2024. PMID: 39040139 Free PMC article.
-
Comparison of topological clustering within protein networks using edge metrics that evaluate full sequence, full structure, and active site microenvironment similarity.Protein Sci. 2015 Sep;24(9):1423-39. doi: 10.1002/pro.2724. Epub 2015 Aug 18. Protein Sci. 2015. PMID: 26073648 Free PMC article.
-
Phylogeographic analysis reveals association of tick-borne pathogen, Anaplasma marginale, MSP1a sequences with ecological traits affecting tick vector performance.BMC Biol. 2009 Sep 1;7:57. doi: 10.1186/1741-7007-7-57. BMC Biol. 2009. PMID: 19723295 Free PMC article.
-
Advanced methods for missing values imputation based on similarity learning.PeerJ Comput Sci. 2021 Jul 21;7:e619. doi: 10.7717/peerj-cs.619. eCollection 2021. PeerJ Comput Sci. 2021. PMID: 34395861 Free PMC article.
Cited by
-
Transovarial Transmission of Anaplasma marginale in Rhipicephalus (Boophilus) microplus Ticks Results in a Bottleneck for Strain Diversity.Pathogens. 2023 Aug 2;12(8):1010. doi: 10.3390/pathogens12081010. Pathogens. 2023. PMID: 37623970 Free PMC article.
-
Sequence Similarity Network Analysis Provides Insight into the Temporal and Geographical Distribution of Mutations in SARS-CoV-2 Spike Protein.Viruses. 2022 Jul 29;14(8):1672. doi: 10.3390/v14081672. Viruses. 2022. PMID: 36016294 Free PMC article.
-
Network analysis of driver genes in human cancers.Front Bioinform. 2024 Jul 8;4:1365200. doi: 10.3389/fbinf.2024.1365200. eCollection 2024. Front Bioinform. 2024. PMID: 39040139 Free PMC article.
-
A tool to enhance antimicrobial stewardship using similarity networks to identify antimicrobial resistance patterns across farms.Sci Rep. 2023 Feb 20;13(1):2931. doi: 10.1038/s41598-023-29980-4. Sci Rep. 2023. PMID: 36804990 Free PMC article.
References
-
- Roberts A, McMillan L, Wang W, Parker J, Rusyn I, Threadgill D. Inferring missing genotypes in large SNP panels using fast nearest-neighbor searches over sliding windows. Bioinformatics. 2007;23(13):401–7. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

