Protein Microarray Analysis of the Specificity and Cross-Reactivity of Influenza Virus Hemagglutinin-Specific Antibodies
- PMID: 30541779
- PMCID: PMC6291623
- DOI: 10.1128/mSphere.00592-18
Protein Microarray Analysis of the Specificity and Cross-Reactivity of Influenza Virus Hemagglutinin-Specific Antibodies
Abstract
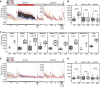
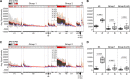
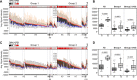
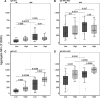
Current seasonal influenza virus vaccines engender antibody-mediated protection that is hemagglutinin (HA) subtype specific and relatively short-lived. Coverage for other subtypes or even variants within a subtype could be improved from a better understanding of the factors that promote HA-specific antibody cross-reactivity. Current assays to evaluate cross-reactivity, such as the ELISA, require a separate test for each antigen and are neither high-throughput nor sample-sparing. To address this need, we produced an array of 283 purified HA proteins from influenza A virus subtypes H1 to H16 and H18 and influenza B virus. To evaluate performance, arrays were probed with sera from individuals before and after a booster dose of inactivated heterologous H5N1 vaccine and naturally infected cases at presentation and follow-up during the 2010 to 2011 influenza season, when H3N2 was prevalent. The response to the H5 vaccine boost was IgG only and confined to H5 variants. The response to natural H3N2 infection consisted of IgG and IgA and was reactive with all H3 variants displayed, as well as against other group 2 HA subtypes. In both groups, responses to HA1 proteins were subtype specific. In contrast, baseline signals were higher, and responses broader, against full-length HA proteins (HA1+HA2) compared to HA1 alone. We propose that these elevated baseline signals and breadth come from the recognition of conserved epitopes in the stalk domain by cross-reactive antibodies accumulated from previous exposure(s) to seasonal influenza virus. This array is a valuable high-throughput alternative to the ELISA for monitoring specificity and cross-reactivity of HA antibodies and has many applications in vaccine development.IMPORTANCE Seasonal influenza is a serious public health problem because the viral infection spreads easily from person to person and because of antigenic drift in neutralizing epitopes. Influenza vaccination is the most effective way to prevent the disease, although challenging because of the constant evolution of influenza virus subtypes. Our high-throughput protein microarrays allow for interrogation of subunit-specific IgG and IgA responses to 283 different HA proteins comprised of HA1 and HA2 domains as well as full-length HA proteins. This provides a tool that allows for novel insights into the response to exposure to influenza virus antigens. Data generated with our technology will enhance our understanding of the factors that improve the strength, breadth, and durability of vaccine-mediated immune responses and develop more effective vaccines.
Keywords: hemagglutinin; influenza; protein microarrays.
Copyright © 2018 Nakajima et al.
Figures

References
-
- Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, Pereira da Silva S, Aungkulanon S, Buchholz U, Widdowson M-A, Bresee JS, Azziz-Baumgartner E, Cheng P-Y, Dawood F, Foppa I, Olsen S, Haber M, Jeffers C, MacIntyre CR, for the Global Seasonal Influenza-Associated Mortality Collaborator Network. 2018. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391:1285–1300. doi:10.1016/S0140-6736(17)33293-2. - DOI - PMC - PubMed
-
- Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, Skepner E, Lewis NS, Spronken MI, Russell CA, Eropkin MY, Hurt AC, Barr IG, de Jong JC, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Smith DJ. 2013. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342:976–979. doi:10.1126/science.1244730. - DOI - PubMed
-
- World Health Organization. 2010. Pandemic (H1N1) 2009—update 112. www.who.int/csr/don/2010_08_06/en/index.html.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

